We're happy to announce that the Personal Genome Project has received its first donated 23andme exome from a participant! As with the genotyping data acquired from direct to consumer testing companies, the PGP also welcomes donations of larger data sets like genomes and exomes. When assigning PGP nicknames (like "PGP1"), we have decided they should go to individuals who have exome or genome data hosted in the PGP -- whether sequenced by us, or donated to us. Thus, hu97DB4A now has the nickname "PGP18"!
What is exome data?
In my earlier post "The Whole Two Yards" I explained that the PGP is interested in whole genomes rather than genotyping. Exome sequencing is a third category of DNA analysis. So what is an exome?
"Exome sequencing" refers to something much like genome sequencing, but limited to the regions in the genome that code for proteins (the "exons" of genes). Proteins perform most of the functions in the cell: from structures to enzymes to relaying signals, proteins are the workhorses of biology. Thus, most known genetic variations that have significant effects are the result of changes within exons -- changes that disrupt the resulting protein coded by a gene. Surprisingly, these regions only account for 1% of the genome! Because of this, there has been some focus on targeted sequencing of only exons -- hopefully getting almost as much useful sequence for a fraction of the cost.
[caption id="attachment_433" align="aligncenter" width="480"]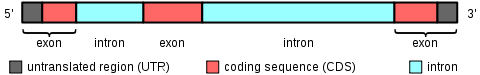 Genes contain protein coding regions ("exons") interspersed with large non-coding gaps ("introns"). To save money, exome sequencing targets and sequences only the exons in the genome -- thereby focusing on the regions most likely to have variations that affect traits. [Image by User:Daycd on en.wikipedia.org, shared as CC-BY-SA][/caption]In the end, isolating exons is difficult, so "exomes" aren't that much cheaper than whole genomes (maybe 2-3x cheaper, not 100x)... but it's still useful to have the cheaper option.1 23andme recently started a pilot exome sequencing service (notably, they provide no interpretation of the data), and some PGP participants have signed up for it.
Genes contain protein coding regions ("exons") interspersed with large non-coding gaps ("introns"). To save money, exome sequencing targets and sequences only the exons in the genome -- thereby focusing on the regions most likely to have variations that affect traits. [Image by User:Daycd on en.wikipedia.org, shared as CC-BY-SA][/caption]In the end, isolating exons is difficult, so "exomes" aren't that much cheaper than whole genomes (maybe 2-3x cheaper, not 100x)... but it's still useful to have the cheaper option.1 23andme recently started a pilot exome sequencing service (notably, they provide no interpretation of the data), and some PGP participants have signed up for it.
Addition of VCF interpretation in GET-Evidence
23andme provided the participant with both a VCF file and individual read data (in the form of a "BAM" file). Personally I'm not a fan of the VCF format for personal genomes, mainly because it fails to report which regions are confidently called as "matching reference". (What this means is that, if a variant isn't listed in the file, you can't tell whether (a) you don't have it, or (b) that region simply wasn't well covered.)
That said, VCF is a very common format, and so I've finally added the ability to interpret VCF to GET-Evidence. I ran the exome data through GET-Evidence and did a little bit of additional interpretation (as with other whole genome reports, these interpretation is far from complete). You can visit the report on GET-Evidence -- and if you'd like a copy of the VCF file itself, it is linked at the top of the report as "source data". We're hoping to reprocess the BAM files to produce higher quality reports and publicly host these larger files as well. For now, though, we're able to immediately accept and interpret VCF files.
Donation of genetic data is very valuable to our project, hopefully we'll see other 23andme exomes donated from participants in the future!
1Exomes have other issues that makes them less desirable, including extremely high variations in coverage, and are difficult to use for detection of larger structural variations (like large deletions or duplications of regions). The PGP does whole genome sequencing because we wish to collect the best data possible, and we feel that a full genome's data is worth the 2-3x higher cost.