Survey about sharing health data online
CLICK HERE TO LEARN MORE AND FILL OUT THE SURVEY !
Funded by the Wellcome Trust on behalf of the Global Alliance for Genomics and Health

CLICK HERE TO LEARN MORE AND FILL OUT THE SURVEY !
Funded by the Wellcome Trust on behalf of the Global Alliance for Genomics and Health

"We do not protect data because the data would take harm; rather, we seek to protect the rights and well-being of individuals who might be harmed by certain uses of their data. This observation could hold the key to protecting personal freedom in a world of evaporating privacy."
Christoph Bock is a principal investigator at the CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences in Vienna. He is also a Project Leader at Genom Austria, a member of the Global Network of Personal Genome Projects. His thoughtful commentary on data protection sheds light on the true dangers of privacy loss, and offers suggestions for how to deal with its potential impact.
Link to article: http://www.nature.com/news/preserve-personal-freedom-in-networked-societies-1.20510
Global PGP Network member PGP-UK (based in London) seeks a postdoc in bioinformatics, statistics or computer science to start Oct 1 and focus on complex trait analysis. They are looking for someone with experience in integrative analysis of multidimensional data, plus a track record of leading a project from conception to publication.
LEARN MORE: http://www.jobs.ac.uk/job/ANV032/research-associate
Sevgi Umur gave a brief update on PGP-UK at the 2016 GET Conference. Watch now.
Recently, my co-worker Abram Connelly scraped the phenotypes in the Harvard Personal Genome Project and made it available in a small SQLite database, publicly available for anyone to download. He made a small webapp around the database where people can play around with the data directly in their browser.
Current webapp (first page has link to gzip of the database) is linked to at the top of this page.
Hello!
I'm Nancy. I recently joined the Harvard Personal Genome Project as a volunteer. I think I've joined at a great time, when the Harvard PGP has the world's largest public dataset that has whole genome sequences linked with genotypes.
I'm excited to join in what I view as an effort that addresses the inherent ethical issues in genomics research: genomes are as individual as a fingerprint, and to stretch the analogy a bit, a smudged fingerprint (de-identified) or summaries of large amounts of fingerprints (aggregation) is only so useful, especially as with the rise of precision medicine we start targeting smaller and smaller subsets of the population with precision medicine.
I think there are many challenges in the HPGP right now, among them challenges in funding and staffing, which contribute to a lot of frustration on behalf of participants, many who have donated blood and saliva samples and waited months and even years without a returned sample from us.
As I've worked with the HPGP staff over the last few months, I've come to see that every last one of the staff members is working extremely hard to get samples sequenced and genomes returned. However, none of us work on HPGP full-time and we also rely on donated effort from other organization, such as sequencing centers (which we're very grateful for!). Although our pace may seem slow, I'm still really impressed by how much work has been done already.
I also like to brainstorm about the future. A future where, among other things, you might be able to check on the status of your genome ala Domino's Pizza instead of having to email us and have to wait for us to laboriously reply to the many emails we get each week.
On that note, happy fall everyone!
Thanks,
--Nancy Ouyang

We're thrilled to announce that data from the Harvard Personal Genome Project is being used in a challenge this year presented by the Critical Assessment of Genome Interpretation (CAGI). CAGI challenges test the ability of researchers to interpret genome data and make phenotype predictions.
PGP data is uniquely valuable for these challenges as it is completely "open source": the algorithms and data can be completely open. In this challenge, experimenters are asked to predict matching phenotype profiles for a set of genomes. To read more about the challenge, follow this link to CAGI's website: https://genomeinterpretation.org/content/4-PGP
Next month, the Harvard Personal Genome Project will hold its annual U.S. conference (MindEx 2015) and labs events (PG-Palooza) in Cambridge, MA. The conference will take place on Saturday, September 12 at Harvard University's famed Sanders Theatre. PG-Palooza labs will be held on Sunday, September 13 at the Cambridge Innovation Center. Thanks to the generosity of our sponsors, all PGP participants will be admitted to both MindEx and PG-Palooza for free!
In years past, the PGP was featured at the GET Conference. This year, the GET Conference is going international. It will take place in Vienna (Sept 17-19, http://www.getconference.org/) and will feature Genom Austria, and other members of the growing international PGP consortium.
For this year's U.S. MindEx conference, the Harvard PGP is working together with the Mind First Foundation, and a focus of the conference will be the mental realm: mind and brain, cognition and behavior. Still, as in previous years, the U.S. conference and labs will provide its established focus on open source genomics and citizen participatory science.
To register as a PGP participant for MindEx, please click here to visit the MindEx and PG-Palooza page at the Harvard PGP website (you'll need to log in to your account), and click on the "Participate" button at the bottom of the page, or go straight to the appropriate EventBrite page (https://mindex.eventbrite.com). We recently made all registration free, so simply use Public Registration. At the conference we'll register you separately for PG-Palooza, which is open only to those enrolled in the PGP.
More about MindEx and PG-Palooza
Conference speakers will include PGP founder and Harvard Professor Dr. George Church, Dr. Ron Kessler (Harvard Medical School), Dr. Martine Rothblatt (United Therapeutics), Dr. Ed Boyden (MIT Synthetic Neurobiology Group), Dr. Richard Wrangham (Harvard), Dr. Madeleine Price Ball (PGP Harvard and Open Humans Project), Dr. Sasha Wait Zaranek (PGP Harvard and Curoverse), Dr. Jordan Smoller (Broad Institute, Harvard Medical, Massachusetts General Hospital), best-selling psychology author David McRaney, gut microbiome experts Justine Debelius and Dr. Siavosh Rezvan Behbahani, and more. PG-Palooza will feature presentations and collections of specimens and data by the Harvard PGP, American Gut, uBiome, LifeNaut, MindModeling@Home, H-Scan, Experiment.com, and more!
For additional details about the conference, labs, speakers, venues, hotels, directions and maps, visit the MindEx conference pages on the Mind First Foundation website (http://mindfirstfoundation.org/mindex2015/).
We hope to see you there!
The following is a guest post by Alan Oppenheimer. The Alan and Priscilla Oppenheimer Foundation seeks to advance humanity through scientific research and education and has been a long-time supporter of the Harvard Personal Genome Project. The views of this guest post, and responses from participants reported upon here, do not necessarily reflect the views of the Harvard Personal Genome Project. It is important to keep in mind that the Harvard Personal Genome Project study is not intended nor expected to help participants diagnose or improve personal health issues.
Following up on our previous blog post, here’s a quick summary of the results of the Harvard Personal Genome Project enrollee survey “What are you looking for in your genome, and how can we help you find it?” There were about 280 respondents.
The first questions were about the participant's background. The “average” participant has been in PGP about 3 years, may or may not have donated a sample, is most interested in inherited disease risk, has 23andMe or FamilyTree/Ancestry DNA data, and is very computer savvy, reading articles/journals.
In terms of the key question in the title of the survey, participants would slightly prefer their genome analysis through either current tools like GET-Evidence and Promethease or an easy-to-use overview tool, versus raw data or a genome browser (see figure below). Primary important factors in exploring their genome include medical analysis and broad, flexible in-depth data, both slightly favored over ease-of-use and accessibility of an overview (and significantly favored over the ability to share/compare with family members).
The most interesting items from the survey were the comments, mainly in the free-form “What else would you like to tell us” question at the end (entered by about 1/3 of the respondents). Most prevalent of those were:
Thanks to everyone who took part. Our next step after this survey: decide on what tool(s) we here at the Oppenheimer Foundation should start building (or assisting the Personal Genome Project with) to best address the survey responses.
[youtube https://www.youtube.com/watch?v=Bo2D4jxAC0U]
At the 2014 Get Conference, Robert Green described how medical genetics is being integrated into primary care, Michael Linderman spoke on how to prepare the next generation of genomicists, and Diana Bianchi presented on how prenatal screening using sequencing of cell-free fetal DNA is revolutionizing prenatal care. Afterwards, they were led in a moderated discussion by Boston Globe reporter Carolyn Johnson. Watch the video.
[youtube https://www.youtube.com/watch?v=RcLhLU2DQss]
If a major goal of genomics research is to understand the underlying molecular causes of beneficial phenotypes, for purposes of promoting overall health in society, then perhaps sports, in many regards, can help facilitate this process. The canonical athletic phenotype, with highly desirable physical traits, may serve as a model for understanding optimal fitness. And certainly professional athletes, at the pinnacle of their respective sport, have tremendous social and economic influence by inspiring everyday athletes and fans alike to emulate their performances. Therefore, a deeper understanding (or at least discussion) of what makes an “elite” athlete, or who has the potential to become one, is warranted. With 99% percent of the human genome being identical, is it plausible to think we all have the inherent ability to become elite athletes? Or, do the remaining 30 million divergent nucleotides of our genetic code determine who can or cannot become an Olympian? At the annual Genes, Environment, and Traits (GET) conference, a sports genomics panel was held to discuss this provocative topic. Invited speakers were:
In a lively debate at the 2014 GET Conference – which included moments of scientific inquiry, levity, and moral contemplation – panelists engaged in discourse over the inheritability and trainability of athletic traits as well as selective pressure from society to enrich for performance phenotypes. Additional topics discussed included:
Practice vs. genetics. Nature vs. nurture. A timeless debate, with a new quantitative spin from current cutting edge advances in next generation genomics technologies. Never before has society had access to such powerful tools to read and write DNA. And athletes, with a history of transcending sport, are as are as popular as ever in mainstream culture. Perhaps the next revolution in science will entail a sports star allowing us all to peak into their biological greatness. Scientists vs. athletes? Why? As the sports genomics panel at the GET conference displayed, these are two communities that stand to benefit from playing on the same team.
(special thanks to moderator Jonathan Scheiman for this written summary)
[youtube https://www.youtube.com/watch?v=eR_wTKwzs9A]
David Altshuler presents his talk, "Future of Genomics" as part of the 2014 GET Conference.
The following is a guest post from Alan and Priscilla Oppenheimer.
If you are enrolled in PGP Harvard, you probably received a recent email that mentioned a survey that we, the Alan & Priscilla Oppenheimer Foundation, are inviting you to take. We'd like to share more about who we are and why we're inviting PGP Harvard participants to take this survey. Although this survey is limited to PGP Harvard participants, we invite others to keep reading. Big changes are ahead that will start affecting us all!
We are a small science-focused family foundation, started in 2007. We knew we were small, but we still wanted to think big. When we became aware of Dr. Church's new Personal Genome Project, we realized that it provided a great opportunity for a foundation like ours to make a big difference. We felt quite privileged when Dr. Church and his team said we could work with them, helping out where we could.
A few of the areas in which we feel we have made a difference include:
Our faith in the PGP in particular and personalized health in general has been validated through a number of recent developments, President Obama's newly announced Precision Medicine initiative being the most visible. Also, as indicated in the recent email, it’s great to see that the PGP has been able to send out almost all submitted enrollee blood samples for sequencing, that the project has spread from Harvard to Canada, the UK, Austria, and beyond, and has spun off important related efforts such as Open Humans.
As the cost of a complete human genome sequence falls towards the $1000 mark, and such sequencing begins to become commonplace, it’s now time to ask the gratifying but difficult question of "What's next?". For the foundation, the answer is related to understanding what our now-obtainable complete sequence means. Helping to address this question has always been an underlying goal of the PGP, but it is only with recent successes that we have been able to begin focusing on it.
The current survey is our attempt to understand the ways in which PGP enrollees (and by extension many others worldwide) want to try to learn about, explore and understand their genomes. With that data in hand we can then focus our limited resources on one or two key tools to aid in that exploration. If you’re enrolled in PGP, we’d thus very much appreciate your taking our 10-minute survey.
Thank you for your time and your interest in personal genomics.
Alan and Priscilla Oppenheimer
The Alan & Priscilla Oppenheimer Foundation
http://www.oppenheimerfoundation.org
Some updates about PGP Harvard: (1) we've added a new feature to the website that allows participants to share their real name, and (2) we have more whole genomes on the way!
The Harvard Personal Genome Project has always emphasized that the genetic data our participants publicly share is "identifiable". This means, even if you remove your name from the data, it's possible for someone to determine your identity. Almost 4,000 people have enrolled knowing that privacy cannot be guaranteed, and many of them are proudly public about their data.
However, to an outside viewer, the data looks anonymous! PGP Harvard's profiles have random identifiers (huID numbers). Even for the staff, we're often unsure whether a participant considers their name to be publicly associated with the profile or not. Sometimes participants do things that seem to indicate they believe their information is public by including their real name in an upload, uploading a photograph, or mentioning their participant ID in another forum. Until now there has been no way for a participant to explicitly choose to associate their name with their data on our website [1].
We'd like the project to look less anonymous and we want to let participants be clear about when they consider their name to be a public fact associated with their data. So we've added to the website a feature that allows a participant to associate their real name. (This is based on their first and last name in our system, which they signed the consent form with.)
To share your real name as a PGP Harvard participant: (1) log in to your account on my.pgp-hms.org, (2) select "Public Profile" from the "Participate" menu, (3) edit the "Real Name" section at the top of this page. Here is a screenshot:
In addition to providing the real names feature to PGP participants, we are also working on processing a new data set received from Complete Genomics, the company responsible for most of the sequencing done by PGP Harvard.
This data comes from around 200 blood samples collected in the past year and a half, including the 2013 GET conference. At this point the most of these genomes have been sequenced and are waiting to be analyzed and approved. We hope to start releasing these to participants soon.
Participants will have a 30- day period to review their data and decide whether or not to withdraw. For everyone that remains a participant, the data will then become public. We look forward to sharing this data and expanding our public resource!
--
[1] There are many participants that have publicly associated their names with their profiles, most notably the first ten participants in PGP Harvard (the "PGP-10"). However, these associations weren't done within the participant website, but were done in other contexts (e.g. conferences, news articles, press releases, blog posts etc).
PGP Harvard is planning two more blood collection events. These events will take place in San Diego, CA on December 16, and in St. Louis, MO on December 29.
PGP Harvard participants who have completed the PGP Participant Survey and all twelve trait surveys are invited to apply to donate blood. Importantly, this event is NOT for those who already have a genome or gave blood at GET2013, GET2014, or at recent Boston or Mountain View collection events.
To apply, please log in to your participant account at my.pgp-hms.org and visit the San Diego collection event page or the St. Louis collection event page. You can complete surveys (or check if you’ve already done them) by visiting the trait surveys page.
We are delighted to announce the launch yesterday of Genom Austria, the fourth member of the Global Network of Personal Genome Projects! This research study is a joint project of the CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences, the Medical University of Vienna, and PersonalGenomes.org. Check out the team.
They launched having already sequenced the whole genomes of two volunteers and plan to enroll and sequence a total of 20 volunteers in the first year. With the addition of Genom Austria, the global network now has member sites at leading institutions in the United States, Canada, United Kingdom and Austria!
Read the press release (PDF).
PersonalGenomes.org is hiring! We are a start-up nonprofit, transforming big ideas about participatory research and open data into resources that can benefit everyone’s health. We are looking for people who are passionate about our mission and excited by the opportunity to work with amazing people all over the globe. We have several open positions, please check them out and share with your family and friends looking for new opportunities:
http://personalgenomes.theresumator.com/apply
PGP Harvard is planning another weekend blood collection event in Boston. The event will take place at Harvard Medical School Saturday, September 20, 10am-4pm.
PGP Harvard participants who have completed the PGP Participant Survey and all twelve trait surveys are invited to apply to donate blood. Importantly, this event is NOT for those who already have a genome or gave blood at GET2013, GET2014, or at recent Boston or Mountain View collection events.
To apply, please log in to your participant account at my.pgp-hms.org and visit the collection event page. You can complete surveys (or check if you’ve already done them) by visiting the trait surveys page.
The following is a copy of our comments as submitted through the online interface at genomicsandhealth.org.
These comments pertain to the International Code of Conduct for Genomic and Health-Related Data Sharing - DRAFT # 6, produced by the Regulatory and Ethics Working Group of the Global Alliance for Genomics and Health. That draft document can be found at this URL: [http://genomicsandhealth.org/our-work/work-products/international-code-conduct-genomic-and-health-related-data-sharing-draft-6](http://genomicsandhealth.org/our-work/work-products/international-code-conduct-genomic-and-health-related-data-sharing-draft-6)
Our most important points are the first two. The first suggests an explicit mandate to inform individuals, families, and communities regarding identifiability of their data. The second suggests individuals, families, and communities from whom data is derived also be considered as potential data sharing recipients.
These comments come from the following Personal Genome Project (PGP)-associated contributors:
To respect individuals, families, and communities, and to foster trust and integrity, we strongly believe the foundational principles should mean that individuals, families, and communities be informed about the identifiability of data relating to them. In particular, participants should be informed of the inherent identifiability of an individual from their genome, or from genotype profiling of multiple loci in their genome. To make this clear, section 4.2 of the guidelines:
4.2 Informing individuals, families and communities about the use and exchange of data relating to them, depending on the nature of the data.
Could be changed to specifically mention identifiability:
4.2 Informing individuals, families and communities about the use and exchange of data relating to them, including its identifiability, depending on the nature of the data.
To respect individuals, families, and communities, and to foster trust and reciprocity, we strongly believe the foundational principles should mean that individuals, families, and communities from whom data are derived also be considered as potential data sharing recipients. To reflect this, section 5.2 of the guidelines could be updated to also describe consideration of the risks of data sharing to/with individuals, families, and communities (in addition to on/about):
5.2 Considering the realistic harms and benefits of data sharing on individuals, families and communities, including opportunity costs.
To also state “with” individuals, families, and communities:
5.2 Considering the realistic harms and benefits of data sharing on and with individuals, families and communities, including opportunity costs associated with both sharing and not sharing.
There are a couple of terms in the draft that have meanings that vary considerably depending on country and legal context. Because this document is intended to convey global policy, we suggest avoiding these terms and, if appropriate, replacing them with terms which avoid unintended or inconsistent legal interpretation.
The first of these is the phrase “moral interests”. One interpretation of this is as “moral rights”, a term that, to our knowledge, varies markedly in its legal meaning. While we recognize the phrase “moral interests” reflects language in Article 27 of the UDHR, we recommend possibly avoiding it to reduce divergent understandings of the meaning of this document.
The other phrase with variable legal meaning is the term “good faith”. As with “moral rights”, in some countries and legal contexts “good faith” has a concrete legal meaning and can be breached. In other contexts, it is an appeal for fair behavior with no legal force.
If not generally binding or enforceable, we suggest changing the phrase:
This code applies to
To state:
This code can potentially be applied to
The third foundational principle refers to what seems like two principles that aren't strongly related: “advancing research” and “fair distribution of [research] benefits”. Also, because genomics research is often not related to health (e.g. ancestry), emphasis on “health and wellbeing” as a principle in themselves (the first principle) could be seen as implicitly excluding these fields of research. We suggest stronger emphasis of “research and scientific knowledge” would be more inclusive. Because “health and wellbeing” seem more related to “fair distribution of benefits”, we suggest rewording the foundational principles from:
1. Promote Health and Wellbeing
2. Respect Individuals, Families and Communities
3. Advance Research and the Fair Distribution of Benefits
4. Foster Trust, Integrity and Reciprocity
To instead be:
1. Advance Research and Scientific Knowledge
2. Respect Individuals, Families and Communities
3. Promote Health, Wellbeing, and Fair Distribution of Benefits
4. Foster Trust, Integrity and Reciprocity
In keeping with the second foundational principle (respect for individuals, families, and communities), we suggest explicitly naming “donors” as those who are giving consent in this sentence:
This Code applies to data that has been consented to for use and/or approved therefor by competent authorities.
To state:
This Code applies to data that has been consented to by donors (or their legal representatives) for use and/or approved therefor by competent authorities.
To enable investigators to ensure that their data has been generated from well-consented sources, we recommend updating the phrase:
...tracking the chain of data exchange.
to state:
...tracking the chain of data exchange to its source.
Because perfect data security is not achievable, we recommend changing the phrase:
Installing strict data security measures to prevent unauthorized access, data loss and misuse....
To state:
Installing strict data security measures to mitigate the risk of unauthorized access, data loss and misuse....
The title for this section, “Minimizing Harm and Maximizing Benefits”, refers two very different extremes in decision-making. To communicate balancing consideration, we recommend changing the phrase and title:
minimizing harm and maximizing benefits
To instead be:
risk-benefit analysis
It was also unclear to us what outcomes would be considered as potential harms or benefits; it might also be helpful to give examples of these.
We've collected blood in Boston before at the GET conference, but attending the event isn't always possible for local residents, so we've decided to hold a blood collection event on a weekend. We're planning a sample collection at Harvard Medical School next Saturday June 21st, 10am-4pm.
PGP Harvard participants who have completed the PGP Participant Survey and all twelve trait surveys are invited to apply to donate blood. Also, this is for folks that aren't already in the sequencing pipeline - no need to attend if you already have a genome or gave blood at GET2013 or GET2014. To apply, please log in to your participant account at my.pgp-hms.org and visit the collection event page. You can complete surveys (or check if you've already done them) by visiting the trait surveys page.
At PGP Harvard our participants are, by and large, very enthusiastic about understanding genetics and their own genomes. Many participants are programmers, researchers, and often both! It should come as no surprise that our staff are often asked “can I see more of the raw data?”
[caption id="attachment_1380" align="alignright" width="300"] Some drives our genomes arrived on. Porsche design! That’s how you know it’s quality.
Some drives our genomes arrived on. Porsche design! That’s how you know it’s quality.
© 2012 Alexander Wait Zaranek, CC-BY license.[/caption]
We’ve always wanted the entire “raw data” to be public, for participants and researchers alike. One issue that stymied us was the intractable size of the data: this sort of data is typically shipped on terabyte disks. I’m now happy to share that we now have an answer and a place to find the data, although accessing this requires some familiarity with using a command line interface and maybe a smidge of programming.
The full data sets PGP Harvard received from Complete Genomics are now shared on a public bucket on Google Cloud Storage, using credits generously donated by Google. Data is organized by huID.
The bucket: gs://pgp-harvard-data-public
To access the bucket, you should read about installing and using gsutil.
Some example commands
List contents of bucket top level:
gsutil ls gs://pgp-harvard-data-public
Recursively list contents of hu011C57 directory, with date and file size details:
gsutil ls -Rl gs://pgp-harvard-data-public/hu011C57
Download/copy the var file from hu011C57 Complete Genomics data to your current directory (234 MB):
gsutil cp gs://pgp-harvard-data-public/hu011C57/GS000018120-DID/GS000015172-ASM/GS01669-DNA_B05/ASM/var-GS000015172-ASM.tsv.bz2 .
With multi-threading and recursion, copy the hu011C57 directory to your current directory. (40.8 GB):
gsutil -m cp -R gs://pgp-harvard-data-public/hu011C57 .
Use a Google Compute Engine VM to analyze the data
You can also access this data using virtual machines in the Google Compute Engine - this could save you a lot of disk space! Once you have a virtual machine you can, for example, use the Python Client Library to automatically access data.
Taking a group photo of Harvard Personal Genome Project participants in attendance at the GET Conference has become a fun annual tradition (2014, 2013, 2012). This year, the group photo was taken on April 29, 2014 at the GET Labs event held at the IBM Innovation Center in Cambridge MA:
[caption id="attachment_1375" align="alignnone" width="600"] Group photo of Harvard Personal Genome Project participants who attended GET Labs on April 29, 2014 in Cambridge MA. Photo credit: Aurelien Dailly for PersonalGenomes.org, CC-BY. We were lucky to have French photographer Aurelien Dailly, who snapped this photo. He is traveling throughout the United States for three months exploring people and places involved in open innovation and DIYbio. Check out his portfolio of photos from his journey thus far.[/caption]
Group photo of Harvard Personal Genome Project participants who attended GET Labs on April 29, 2014 in Cambridge MA. Photo credit: Aurelien Dailly for PersonalGenomes.org, CC-BY. We were lucky to have French photographer Aurelien Dailly, who snapped this photo. He is traveling throughout the United States for three months exploring people and places involved in open innovation and DIYbio. Check out his portfolio of photos from his journey thus far.[/caption]
[caption id="attachment_1352" align="alignright" width="108"] Blood samples in EDTA tubes, CC-BY-SA, by Lennart B.[/caption]
Blood samples in EDTA tubes, CC-BY-SA, by Lennart B.[/caption]
Blood is our current best source for getting DNA for whole genome sequencing. For PGP Harvard, GET conference blood collections in Boston have been a great success. But we know not all participants can travel to Boston for these events, so we want to pilot blood collection events in other cities. Our next event will be in Mountain View next week on Wednesday May 7th, between 1pm-5pm.
PGP Harvard participants who have completed the PGP Participant Survey and all twelve trait surveys are invited to apply to donate blood. To apply, please log in to your participant account at my.pgp-hms.org and visit the collection event page. You can complete surveys (or check if you've already done them) by visiting the trait surveys page.
Tomorrow we are bringing together over 100 Harvard Personal Genome Project participants and 20 research groups who wish to collaborate with them and make some science together! We're thrilled that the New York Times is featuring a profile of the event and its attendees (i.e. "omic astronauts") in tomorrow's print edition. Check it out!
This is the fifth year that the nonprofit PersonalGenomes.org has organized the GET Conference, and it is going to be the best year yet. One really exciting aspect is GET Labs, which we made into a standalone event the day before the regular conference. The focus is on *doing* science, not talking about it. We bring together a cohort of extremely well-characterized and well-consented individuals enrolled in the Harvard Personal Genome Project & researchers who wish to study them. Around 20 research groups signed-up to attend this year, and will be performing a wide range of activities from armpit microbiomes to adult stem cell establishment. Everyone gets a little passport for documenting their adventures in health research.
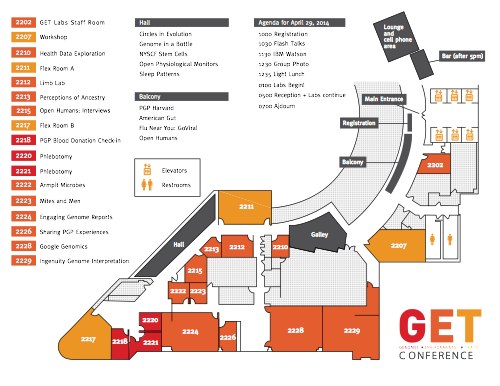
Agenda and map of GET Labs, seea high-resolution PDF.
You can read more about the research groups participating in GET Labs, here:
GET Labs and GET Conference next week are going to be a blast! We had a really cool idea I'm going to be excited to see in action: GET Labs passports!
Jason got some little notebooks for attendees to use during the day, with the plan of stamping the front with a GET Conference stamp:
Then Mike Chou suggested: why not a stamp for every activity? Then participants can collect stamps from each activity through the day! So we designed those and they just arrived - and they look awesome:
 Check out the GET conference site to learn more about the participants and researchers that will attend GET Labs!
Check out the GET conference site to learn more about the participants and researchers that will attend GET Labs!
For many of you this notice is a repeat – we've opened up our application for Harvard PGP participants to attend GET Labs! This year GET Labs will be its own event on April 29th, at the IBM Innovation Center in Cambridge, MA. (The GET conference is the following day, and will have separate tickets.) We have over a dozen labs already signed up to work with participants – and planning to return resulting research data! Participants will also be welcome to organize their own sessions and gatherings.
Space is limited and to ensure that our researchers find GET Labs a valuable experience, we've limited our invitations to Harvard PGP participants that have shared significant genetic data on their public profile (genome, exome, or genome-scale genotyping like 23andme or Family Tree DNA) and have completed the current set of surveys.
If you're not a participant but have genome-scale data you want to share publicly from 23andme or Family Tree DNA, you can enroll now in the Harvard PGP – we'll accept new enrollees as GET Labs participants, provided they have this data shared publicly.
I'm happy to announce the publication of our paper "Harvard Personal Genome Project: lessons from participatory research" in Genome Medicine, a general retrospective of the current Harvard Personal Genome Project. There is growing interest in participatory research and data sharing (with either participants or researchers), and the Harvard PGP has concrete experience relevant to these conversations. In this paper we share our experiences with the hope of encouraging and assisting others interested in similar research models.
In the rest of the post I’ll give some personal summary, written colloquially. Although many observations seem obvious or trivial, remember that hindsight is 20/20! This wasn’t always what people expected, and that’s why it’s important to share our experiences.
What happens when your project gives participants data access?
One thing that doesn’t happen (at least, not yet): the sky doesn’t fall. Our report explores this question in the form of quantifying the communications we receive from participants. Only one in ten participants that received access to whole genome data through the PGP followed up with us asking any questions about that data – and the majority of those questions were regarding file formats and additional files, not interpretation.1 For a project whose public image is "Personal Genomes!" (and whose participants are extremely engaged and interested in genomes), I think this is a very modest amount of "customer support". This should be reassuring to any project considering providing genetic data access to its participants.
One thing that does happen: when you tell participants you plan to give them data, they wonder where the data is. Over a quarter of our communications are from participants eager to be sampled and receive data. Research timelines are slow. Sometimes analyses fail. It’s really hard to tell someone "Your sample from 14 months ago is still sitting in the freezer".2 Participants are used to medical tests and commercial products which return results in days or weeks. But those tests are fast because commercial/clinical providers have developed standard, polished processes. Researchers have a much fuzzier idea of where they're going – that’s why it’s research! – and things almost never go as smoothly as the researcher imagines they will.3
Unexpected positive consequences of public participatory research
What happens when you have a public project and don’t promise anonymity? Participants talk to each other! They create forums, they write books, they report on their experiences. They can even complain about you – a powerful empowerment of participants.
But researchers, don't shy away from this scary new world where participants become people. There are some significant benefits for your work when participants become part of your community. An engaged cohort wants to be studied! They go out of their way to give you samples. They give you data. They find errors. They perform follow-up investigations and tell you what happened. You get participants who don't merely expect you to study them, they study themselves.
1: Regarding that interpretation – our interpretation process is semi-automatic and creates fairly limited, highly technical summaries (these genome reports are publicly shared on GET-Evidence).
2: Part of why it's hard is because the answers are often embarrassing, e.g. "Alexandra started that project, but she moved to Tokyo and nobody can read her notes". (This is hypothetical! I think the Harvard PGP has done much better than this! But it's not an unusual story.)
3: Researchers are also generally optimistic about timelines. If the researcher says it should take two weeks, expect it to take six. Maybe ten. If you think about it, it's just human nature – to want to do the research in the first place, the researcher must be excited and optimistic about it!
Quick note: Tomorrow Jeantine Lunshof, Barbara Prainsack, and John Wilbanks will have a live chat ("Do You Have a Right to Your Personal Data?") hosted by Science's senior commentary editor, Brad Wible. You can join them at 3pm EST tomorrow for a live chat. (You can also leave comments or questions on that page, ahead of the chat.) Also, if you haven't seen it already, their Science policy piece is publicly accessible ("Raw Personal Data: Providing Access", by Lunshof, Church, and Prainsack).
I'm thrilled to report the publication in Science today of "Raw Personal Data: Providing Access", a Policy Forum opinion piece by Jeantine Lunshof, George Church, and Barbara Prainsack. As the ethics advisor to the Personal Genome Project, Jeantine, along with George, has pioneered our "open consent" model for public data sharing. With this piece, Lunshof et al. address the topic of sharing data with participants themselves. They advocate that participants deserve access to raw data in any research and clinical setting, to establish fairness and reciprocity in research and any other context where data are handed over.
This vision is very much in line with the Personal Genome Project's work since 2005, and more recently with our public comments on the NIH draft Genomic Data Sharing Policy last November and our recently funded Open Humans Network. Data relevant to understanding human health and biology is often very personal, very identifiable, and – as a result – difficult to share. Participant-mediated data sharing is a way to enable open human data.
As Lunshof, Church, and Prainsack point out, modern information technology should facilitate data sharing with participants. It is understandable that such a model has been impractical in the past, but the technology is now ready. To make it a reality, we need to see our expectations change regarding how research studies should operate.
Lunshof et al. make a careful distinction between "access to data" and "return of findings" – the first is the raw research data alone, while the latter involves interpretation of that data (a process sometimes requiring instrument certification or clinical expertise). Their piece challenges researchers to treat access to raw research data as a separate issue from its interpretation; they call for researchers to grant raw data access and thereby promote participant agency, establishing a more fair and reciprocal relationship.
We at PersonalGenomes.org are thrilled to announce that our Open Humans Network was awarded a $500,000 grant from the Knight News Challenge: Health. The winners were announced at the Clinton Foundation Health Matters Conference on January 14, 2014 in La Quinta, CA.
The John S. and James L. Knight Foundation is the leading funder of journalism and media innovation. It has been seeking the next generation of innovations to inform and engage communities.

With its Knight News Challenge: Health, the Knight Foundation is funding breakthrough ideas that harness data and information for the health of communities. The five-stage competition began in August 2013 with an “Inspiration Phase” during which anyone could post needs, interests, and ideas online at its website, and continued with the submission of 630 health and data news projects, all competing for a share of $2.2 million in funding and support. After a feedback stage, 39 semi-finalists – the Open Humans Network among them—were invited to refine their projects and submit videos prior to judging. Seven winners were awarded grants.
The Open Humans Network, led by myself and Madeleine Ball of PersonalGenomes.org, attempts to break down health data silos through an online portal that will connect participants willing to share data about themselves publicly with researchers who are interested in using that public data and contributing their analyses and insight to it. The portal will showcase public health data and facilitate its exploration and download. The Open Humans Network ultimately hopes to revolutionize research by making it easy for anyone to participate in research projects and facilitating highly integrated, longitudinal health data. This portal will consist of three components: individual data profile pages, a public data explorer and a set of design guidelines for researchers seeking a collaborative data-sharing model.
The Open Humans Network grows out of the Personal Genome Project (PGP), a research study founded in 2005 that has pioneered open-access sharing of genomic and trait data. Through our years of work on the PGP, we recognized the need to link together the people and data from many exciting open research efforts.
Current partners with the Open Humans Network include the Harvard Personal Genome Project (PI: George Church, Harvard Medical School), American Gut project (PI: Rob Knight, University of Colorado, Boulder), Flu Near You: GoViral Study (Rumi Chunara, Boston Children’s Hospital) and the Mt. Sinai School of Medicine (Eric Schadt, Icahn Institute). By helping participants locate legitimate, open research projects and promoting data sharing, the Open Humans Network will allow any individual to make valuable contributions to science.
We believe that everyone benefits from a health research community that is more transparent, efficient, and equitable. Toward this end, we aim to reimagine health research and biodiscovery! Our sleeves are already rolled up, and we are tremendously excited to have the support from the John S. and James L. Knight Foundation’s Knight News Challenge to help us make our vision a reality.
For more information, visit us at: http://openhumans.org
Jason Bobe
Executive Director
PersonalGenomes.org
-- 501(c)(3) nonprofit organization
Rumi Chunara is an Instructor and Researcher working with the group HealthMap at Harvard Medical School and Boston Children’s Hospital. She brings together her background in engineering and epidemiology to build and use new tools for improving public health and our knowledge of how and why diseases spread. GoViral is the newest endeavor; a platform that creates a data-driven view of public health that the public can participate in and use directly.

What is Flu Near You and the GoViral study?
Flu Near You is a free and real-time online flu surveillance system administered by Healthmap, a research group at Boston Children’s Hospital. The Flu Near You website and mobile application allows the public to report their health information using a quick weekly survey. Using participant-reported symptoms, Flu Near You graphs and maps this information to provide local and national views of influenza-like illness.
This year we are recruiting people from Massachusetts for GoViral; a next-generation study that will revolutionize infectious disease prevention and awareness by making it community-based and data-driven.
GoViral combines for the first time, online crowdsourced data and diagnostic samples that people can provide themselves from their homes.
What will GoViral participants do?
Participants will receive a weekly survey (via email or push notification on our smartphone app) that takes about 20 seconds to fill out, asking which of 10 symptoms you had in the previous week. Participants will also receive a GoViral test kit they can keep at home. If at anytime you get sick with a fever and sore throat or cough, you will be asked to provide a really easy to obtain spit and/or nasal-swab sample. Participants will also receive a Do-It-Yourself home test that detects 4 different kinds of flu and cold viruses, and all samples will be analyzed at a central laboratory that checks for 20 different viral infections. The kits can be returned by USPS and results from these tests will be used for research purposes only.
We will be collecting and testing samples throughout the flu season (until about April). At the end of the season will ask participants to fill out a short survey (2 minutes) about how the information affected their behavior. Flu Near You is completely free to use. Data from each group will be aggregated and visualized in an easy to interpret yet anonymized manner.
GoViral Participants will be able to track their own history throughout the season and as well, through the aggregate data, be able to see in real-time what infections and symptoms are going around right near them so they can take appropriate public health measures and understand when something might be abnormal.
How can PGP volunteers specially contribute or benefit from the GoViral project?
Influenza happens every year and no groups are exempt! There are still a lot of open questions including how many people actually get the flu or another type of respiratory infection. This platform enables us to learn more without relying on who goes in to see the doctor, as not all people do. As well, we believe this type of platform can help keep people healthy by providing them with very relevant information.
We think that PGP volunteers would be great participants because they are excited about and knowledgeable about the valuable information that individuals can hold in regards to our health. In the future it would be interesting to examine genetic makeup and predisposition for respiratory infections!
For the first year of the study we are only recruiting people in Massachusetts, but in subsequent years we can expand to other locations around the country.
Learn more and join here: http://flunearyou.org/goviral
--
NOTE: Enrolled Harvard PGP participants may find more information about this and other studies inside the third party activities page.
The New York Stem Cell Foundation (NYSCF) announced today a collaboration with the Harvard Personal Genome Project (PGP). Initially, we will be seeking out 50 Harvard PGP participants to donate skin samples for generation of induced pluripotent stem cell lines.
Induced pluripotent stem (iPS) cells are derived from adult skin cells and can give rise to any cell type within the human body. This makes them a promising research focus for improving human health. The NYSCF's robotic methods create standardized cells, which is important because it helps scientists understand when cell features are due to genetic or epigenetic differences (and not to the different techniques used to create the cell lines!). PGP participants are a great resource for scientists because their genome and other biological data can be shared publicly. By working together, the NYSCF and PGP will create standard iPS cell lines whose data can be published publicly – a great resource for the scientific community.
Harvard PGP participants interested in working with the NYSCF can view more details about this study on our third party studies page.
Genomics England is the company that will be sequencing the United Kingdom 100K Genomes Project. In response to a question raised during a recent "Town Hall" event, they stated that participants will have access to their data:
Q: Can I have access to my data? And how soon?
A: A patient can have access to their data if they wish and this can be provided to them in the appropriate format. The patient will receive the feedback from the sequencing and analysis of their genome via the clinician who is providing them with on-going care for their disease or condition.
We hope this means that participants will have access to the same "raw data" about their genomes that researchers will. If so, this represents an excellent step forward for both participants and researchers.
In this survey of GWAS studies by Ramoni et al., 4% of studies surveyed had returned individual results to participants. An NIH policy mandating data access for participants, as we recommended last week, would greatly improve this statistic. We hope providing participants access to their personal and identifiable study data becomes the norm rather than the exception.
Yesterday (11/20/13) we submitted to the United States National Institutes of Health (NIH) our public comments on their draft Genomic Data Sharing Policy. This policy will impact numerous participants, mandating the sharing of genetic data – data we know to be identifiable and meaningful. Please read our recommendations below, tell the NIH if you have similar concerns, and share this with others.
The Personal Genome Project (PGP) is a global network of research studies with thousands of participants dedicated to the creation of public resources composed of genome and phenotype data. The first PGP research study was founded at Harvard Medical School in 2005, and international sites now exist in three additional countries.1 The PGP has been at the forefront of participatory research in genome sequencing and has extensive experience with the ethical, privacy, and consent issues involved. We welcome this opportunity to publicly comment on the NIH draft Genomic Data Sharing (GDS) Policy and make recommendations for improvements.
Our recommendations can be summarized as two areas for improvements in section IV.C. of the draft policy: (1) to adequately inform researchers and participants of the inherent identifiability of genetic data, and (2) to require researchers share with participants their personal research data to establish reciprocity and to increase data sharing.
The draft GDS Policy makes no mention of the inherent identifiability of genetic data. All genetic and phenotype data shared is mandated to be "de-identified". Footnote eight of the draft states: “'De-identified' refers to removing information that could be used to associate a dataset or record with a human individual. Under this Policy, data should be de-identified according to the standards set forth in the HHS Regulations for the Protection of Human Subjects and the Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule.”
This definition of "de-identified" is inconsistent: genetic data is inherently identifiable. Using nothing more than genetic data and other publicly available data, researchers were able to identify nearly 50 individuals whose samples were "de-identified" (i.e. all public data met the same standards mandated by this draft).2 It is now a documented fact that this type of genetic data, even if scrubbed of personal information as described in this draft, “could be used to associate a dataset or record with a human individual”. Genetic data itself violates the draft's definition of "de-identified".
In the past, de-identification of samples or data sets by stripping personal data (name, social security number, date of birth, etc.) was sufficient to avoid re-identification of a particular subject. Genetic data was not seen as an equivalently identifiable piece of information. This is demonstrated to no longer be the case, and the identifiability of genetic data is likely to increase and may eventually become trivial. Ancestry databases currently link genetic elements to surname and in the future are likely to link genetic elements to individual ancestors. Controlled-access databases create a legal barrier to re-identification, but data security breaches are possible and have been an increasingly high profile issue in recent years. If the NIH is to mandate that all participants in NIH-funded studies producing large-scale genetic data agree to broad sharing of their genetic and phenotypic data, it is mandating an exposure of many participants to a known re-identification risk.
If the NIH wishes to uphold the public trust in biomedical research, it must respect the right of research participants to be informed of relevant risks. If all potential participants in these studies are asked to agree that their “genomic and phenotypic data may be shared broadly for future research use” (link), they must also be adequately informed regarding the identifiability of that data.
We recommend this draft be amended to:
The draft GDS Policy mandates all NIH-funded research studies that wish to produce "large-scale"3 human genetic data require that all participants from whom samples are collected consent that their “genomic and phenotypic data may be shared broadly for future research use” (link). This is elsewhere defined as NIH-designated controlled-access or open-access databases (the latter only if participants “have provided explicit consent for sharing their data through open-access mechanisms”).
What is not addressed in this draft is a statement about genomic data sharing with the participants themselves. We strongly recommend the NIH consider including such a requirement for two reasons.
The first reason is to establish reciprocity in the data sharing mandate. This draft mandates all participants in NIH-funded studies generating large-scale genetic data allow broad access to their genomic and phenotypic data to unknown individuals – without ever having access to that data themselves. Participants' genetic data is sensitive, meaningful, and identifiable. Participants deserve the reciprocal mandate that their personal data being shared with others also be shared with them.
The second reason is that this is a significant opportunity to further the NIH's data sharing goals. Participant-managed data sharing is a promising mechanism for open-access data sharing. Even if participants would not have agreed to open-access at the outset of a study, their attitudes may change. Additionally, participants may wish to share their data with future studies in a selective manner. Participant access to data enables an additional participant-managed model for data sharing, and we can imagine a future where numerous studies benefit from participant-donated data.
We recommend the following:
1) In section IV.C.4: “If there are compelling scientific reasons that necessitate the use of cell lines or clinical specimens that were created or collected after the effective date of this Policy and that lack consent for research use and data sharing, investigators should provide a justification for the use of any such materials in the funding request.” We suggest clarification of whether the lack of informed consent automatically exempts the researcher from data sharing, or if data sharing is expected to occur despite the exemption.
2) We suggest clarification confirming that “sample identification” using genomic data or other genotypic assays which are not intended to identify individual human participants is acceptable (e.g. detection of duplicate samples across different studies for statistical validity or for quality assurance).
3) “Binary alignment matrix (BAM)” should probably be "Binary Alignment/Map (BAM)". Assuming this is a reference to SAM and BAM files, there is no clear definition what the BAM acronym abbreviates ("B" could potentially mean "BGZF" or "Binary"), but a SAM file is defined here as a "Sequence Alignment/Map": http://samtools.sourceforge.net/SAMv1.pdf
Many thanks to the Harvard PGP staff that contributed to these recommendations: Madeleine Ball, Jason Bobe, Michael Chou, George Church, Tom Clegg, Preston Estep, Jeantine Lunshof, and Alexander Wait Zaranek
[1] Three PGP sites exist currently outside the United States: (1) PGP-Canada, based out of the McLaughlin Centre, University Toronto & Sick Kids Hospital (2) PGP-UK, based out of the University College London and (3) another site in the EU with ethics approval, set to launch in early 2014. The PGP Global network is coordinated by PersonalGenomes.org, a 501(c)(3) nonprofit based in Boston, Massachusetts. To learn more please visit: http://www.personalgenomes.org/mission
[2] Gymrek M, McGuire AL, Golan D, Halperin E, Erlich Y. "Identifying personal genomes by surname inference." Science. 2013 Jan 18;339(6117):321-4.
[3] Defined as more than 100 participants for genotyping or multi-gene sequence data, or whole genome sequence from a single participant.
Link to PDF version of these public comments: NIH_PGP_Public_Comments_GDS_Policy_11202013.pdf
[caption id="attachment_1225" align="alignright" width="300"] Union flag. Photo credit: Thanks to Flickr user jsteph, cc-by-nc-nd.[/caption]
Union flag. Photo credit: Thanks to Flickr user jsteph, cc-by-nc-nd.[/caption]
We are pleased to announce the launch of the Personal Genome Project UK (PGP-UK), which is the third site in our global network and the first in Europe! The PGP-UK team is composed of Stephan Beck (University College London), Jane Kaye (University of Oxford), Rifat Hamoudi (University College London) and a great groups of advisors.
Nearly 500 people had written to PersonalGenomes.org over the past few years requesting that we launch a PGP research study for residents of the United Kindom. Until today the PGP was available only to people in the USA (Harvard Medical School) and Canada (McLaughlin Centre, University of Toronto). We're working on growing the global PGP network to include dozens of countries where individuals and researchers are interested in working to create public data resources.
A press briefing was held at the Wellcome Trust yesterday that included:
A summary of press so far:
This weekend we switched www.personalgenomes.org to a nice new site. The new design is cleaner and hopefully easier to use.
It also features a redesign which represents the Harvard Personal Genome Project as one of multiple PGP sites. The Harvard PGP has been a pilot site and is still the origin of almost all current PGP public data and cell lines, but our project only enrolls volunteers with United States citizenship or permanent residency. The world is larger than that! Right now the only other site is the Canadian PGP site, but PersonalGenomes.Org looks forward to seeing to many other PGP sites around the world.
We hope to add more content and features to the site in coming weeks. I did almost all of the code/design portions, using Bootstrap 2 and Django. Let us know if you find any bugs.
 Abigail Wark is a research fellow in the Tabin Laboratory in the Department of Genetics at Harvard Medical School. Her research focuses on understanding the causes and consequences of variation in uniquely human traits. She is the Project Director for Circles in Human Evolution: the Areola, a citizen science collaboration with the PGP that is gearing up to provide the worlds first genetic study of diversity in human areolas.
Abigail Wark is a research fellow in the Tabin Laboratory in the Department of Genetics at Harvard Medical School. Her research focuses on understanding the causes and consequences of variation in uniquely human traits. She is the Project Director for Circles in Human Evolution: the Areola, a citizen science collaboration with the PGP that is gearing up to provide the worlds first genetic study of diversity in human areolas.
PersonalGenomes.org has partnered with the Tabin Laboratory and Circles in Human Evolution to create a third party research opportunity for PGP volunteers. Abby recently sat down with us to answer questions about this project.
Why study the genetics of human diversity?
We are living in the golden age of human genetics. The bulk of what we know about genetics so far has to do with genes that have gone awry. Focusing on genetic dysfunction makes sense because we want to prevent and cure human disease. But strongly detrimental genetic variants are a very small part of human genetic diversity. The truth is that most of the variants we carry in our genomes do not cause us catastrophic problems. These variants help make each of us who we are; they make us biologically unique. But while we have learned so much about genetic origins of disease, we know almost nothing about the genetic signature of healthy diversity. The tools are available now to update our view on this, to examine and understand a bigger slice of human biodiversity.
Are there meaningful consequences to this genetic diversity?
Of course! Nature is full of examples where genetic diversity can have real functional significance. Sensory systems provide dramatic examples of this because they can lead animals to interpret the world in completely different ways. For example, think of an insect with a gene that enables it to see UV light. That insect has access to information that you and I don’t have. The same thing is true for much more subtle cases of diversity. If you are a fish, subtle changes to your sensory systems can affect your likelihood to school, which has big ramifications for how you live your life as a fish, how your respond to predators, etc. I think we all know intuitively that this is true for humans too. We vary in all kinds of traits, from physical traits like height and hair color to sensory traits like taste or odor perception. And there’s reason to believe that these traits can impact how we live our lives, from what foods we eat to how well we deal with hot weather. The era of personal genomics offers the chance to understand what this diversity is made of and what role it plays in our lives.
Why did you choose to study the areola?
Areolas probably seem like such a strange topic! But they turn out to be pretty fascinating. Areolas are the circular markings that surround the human nipple. Did you know that no other animals have these spots? They are defining marks of our species! The fact that areolas are circular is really significant for this study. Circles are one of nature’s simplest forms. Many interesting human traits are the opposite of this – they are extremely complex. The genetic recipe for building a human hand or brain is complicated which makes it very challenging to draw a line from even the simplest genetic changes to effects on human traits. But circles are simple to build and simple to change. It turns out that the simplicity of circles might give us a foothold for discovering the genetics pathways that define human traits.
So that is why areolas are a practical, though somewhat unusual topic, for studying human genetics. That said, one thing that has made this project really fascinating for us is that areolas are much more than just circular markings. In women, the pigmentation of the areola is believed to signal fertility and some evidence suggests that this may be one of the first indications of pregnancy. Areolas play an important role in nursing, providing both a visible target and a pheromonal attractant for newborn infants. In fact, the number of areolar glands, which differs from person to person, has been associated with infant weight gain. Developmentally, these glands are related to the mammary glands and one of the really exciting angles of our work is to see whether the areola might provide a window into the inner biology of breast. If it does, the results could be really important for many aspects of breast health, from lactation to cancer.
Why are you using a citizen science approach? What role do participants play?
Citizen-science is a movement to give non-scientists an active role in scientific discovery. I like to think that we are all potential scientists, just with different amounts of training. As children, many of us took delight in the act of discovery. We explored our backyards, tried to grow plants, observed animals, and mixed toothpaste and ketchup to see what we would get. As I see it, some of those kids became scientists and most did not, but there’s no reason to exclude non-scientists from that sense of discovery. In our project, participants become part of our field research team. They are given the tools and information they need to make observations about their own bodies. They share that data with us and our job is to compile and analyze the findings for the whole community. The whole project is a partnership.
Can participants sign up?
Yes, please do sign up! This is a PGP-specific study, so only PGP participants can join. This limits the number of people who can help, so we need you and all of your PGP friends! I want to emphasize that both men and women are encouraged to join our study. Submitting photographs through our server is very helpful, but is completely optional. Your data is still useful to us even if you choose not to include photographs.
To read more about the study or to sign up, please go to: https://my.personalgenomes.org/third_party/12
[caption id="attachment_1177" align="alignleft" width="600"] PGP participants at the 2013 GET Conference. Photo credit: PersonalGenomes.org, license CC-BY[/caption]
PGP participants at the 2013 GET Conference. Photo credit: PersonalGenomes.org, license CC-BY[/caption]
Around 150 Personal Genome Project (PGP) participants attended the 2013 GET Conference and it has become a tradition to take a group photo. Here is the group photo from last year's GET Conference.
The data and samples that participants share in the Personal Genome Project (PGP) are considered highly identifiable. One of the key aspects for defining what it means to be an implementation of the Personal Genome Project is an absence of anonymity:
From our guidelines for PGP implementations:
"Non-anonymous. The risks of participant re-identification are addressed up front, as an integral part of the consent and enrollment process; neither anonymity nor confidentiality of participant identities or their data are promised to research participants."
We have designed a consent process that includes many layers of upfront and ongoing education about the unique nature of public genomics research studies like the PGP. One of the important messages to participants is that their data are highly identifiable and therefore not “anonymous”. For example, the study guide that accompanies our mandatory entrance exam provides one of the more famous examples of how only a few pieces of demographic data can reveal a person’s identity:
From the PGP Study Guide:
"Identities can be discovered with surprisingly little information -- for example, the combination of sex, birth date and ZIP code is specific enough to be uniquely identifying information for 87% of people!"
We know that hands-on demonstrations of otherwise abstract concepts can be extremely valuable for learning. Talking about a “personal genome” in the abstract can be a far different experience compared to wading through millions of variants contained in your very own personal genome sequence! So to enhance understanding of identifiability, we invited two research groups to demonstrate how re-identification is possible using public PGP data during GET Labs in Boston (April 25-26).
Latanya Sweeney’s Data Privacy Lab drew upon her pioneering work on the identifiability of demographic data to show how these techniques can be applied to public PGP profiles containing sex, birth date, and ZIP code. It was no surprise to find that many PGP participants are, in fact, identifiable. Indeed, all PGP participants should expect this potential outcome.
This is important, considering Harvard PGP participants are able to add ZIP codes to their public profiles in anticipation of research activities that explore how geographic location -- and all the associated chemical exposures, microbes, viruses, air quality, allergens, etc. -- impacts health. For anyone who was not at the GET Conference, Sweeney's group has created a tool showing how identifiable you are in your own zip code. Check it out here: http://aboutmyinfo.org/
A word of caution is required here about the best way for PGP participants to respond: we strongly advise any participant concerned about the identifiability of their data to reconsider their participation in the Personal Genome Project. Another viewpoint, one that we find worrisome, is for participants in the PGP to deploy clever tricks for reducing the identifiability of their public data. As part of their demonstration, the Data Privacy Lab is providing tools to participants that “scrub” their data (e.g. replacing a 5 digit zip code with a 3 digit zip code, etc). This may create the impression of privacy, but it will not make participants anonymous. Earlier this year an exciting study published by Gymrek et al. in Yaniv Erlich’s lab forcefully demonstrated that genome data alone is extremely identifying. Melissa Gymrek also had a table at the GET Conference this year where she demonstrated the technique to participants. Their research matched whole genome Y-chromosome data to ancestry databases, which link surnames with Y-chromosome markers. With these surname clues and just a few other pieces of publicly available data, their group was able to identify specific individuals and families from their highly distributed “anonymous” cell lines.
Thus, all participants should believe that they are identifiable: there is no such thing as an “anonymous” genome!
In our experience, many participants want to be identified and are very open about which public profile is theirs. The PGP does not require participants to reveal their names, but with the media coverage of the Sweeney group's work we realize that the project appears to outsiders as “anonymous” — even though participants, after passing our enrollment exam, know better (or should)! To meet the desires of some participants and to further clarify the non-anonymous nature of the PGP, we’re going to work on allowing participants to add their photos and/or name to their public PGP profiles. I expect it will make PGP profile pages much more “personal” and create a provocatively different scientific database!
The (sold out!) 2013 Genomes, Environments and Traits (GET) Conference is taking place this Thursday and Friday in Boston. We are celebrating the 60th anniversary of the DNA double helix with an amazing line-up of speakers and Labs.
You may watch the live webcast for free via our new channel at Fora.TV: get2013.fora.tv. Big thanks to our sponsor, Illumina, for helping to make possible the live streaming.
The hashtag for this event is: #get2013
A couple weeks ago scientists in Europe announced that they had sequenced the genome for the most famous cell line in science: HeLa. The first cell line ever created, HeLa cells have also been the most frequently used: from testing the polio vaccine in the 1950s to current tests of drugs and compounds, these cells are part of the foundation of modern molecular biology. HeLa cells are also a story of communication failures between researchers and the "human subjects" whose samples are studied. Henrietta Lacks -- after whom the cells were named -- was a black tobacco farmer who died in 1951; her tumor tissue was taken and grown without her knowledge or consent. It was decades before her family learned about the cells, and they were frustrated by paternalistic treatment and being kept in ignorance. The story is told in excellent detail in Rebecca Skloot's book, "The Immortal Life of Henrietta Lacks."
[caption id="attachment_1088" align="alignright" width="300"] HeLa cells with fluorescent stain.
HeLa cells with fluorescent stain.
CC0 from Wikipedia.[/caption]
With such a history, we might expect extra sensitivity when researchers perform major research on this cell line. Unfortunately, this was not the case: the researchers had not contacted the Lacks family. That faux pas sparked sharp criticism from Yaniv Erlich, Jonathan Eisen, and Rebecca Skloot. A widespread outcry arose, demanding more respect for the human subjects from whom cell lines are derived. And yet we should understand the researchers didn't do something particularly special: Joe Pickrell pointed out that HeLa's genetic information had been published for some time through projects like ENCODE.
We are noticing the elephant in the room: cell lines were collected from individuals before the modern era of whole genome sequencing. Even if some consent was acquired, little or no warnings were made regarding potential loss of privacy.
And this is where researchers throw up their hands: "Are we supposed to stop our research? Science would come to a halt if we expect this level of consent before publishing sequence data!" The research by Gymrek et al. in Yaniv Erlich's lab comes as an unwelcome confirmation of what George Church has been warning for years: genomes are identifiable and revealing. We may not be able to hide from this much longer: with direct-to-consumer genotyping services, research subjects will theoretically be able to detect when public sequence data matches their private genotyping data.
Specimens can no longer be treated as anonymous, they come from people. This is the new reality and we need to do better.
The Personal Genome Project has been working to address this issue since its inception in 2005. Using an "open consent" process, we collect samples and data from volunteers who have made an informed decision to assume the open-ended risks associated with donating these items to be publicly available. We have a dozen PGP cell lines shared at Coriell, and there's over a hundred more that will be listed soon. When the project started years ago, many wondered how many people would volunteer for the risks of privacy loss. It turns out this fear was probably exaggerated: as I write this we have over 2500 volunteers enrolled.
But we need help, especially from fellow researchers. Here's ways you can help us create a new generation of well-consented samples and data:
[caption id="attachment_1111" align="alignright" width="240"] License:CC-BY, by Brenda Gottsabend[/caption]
License:CC-BY, by Brenda Gottsabend[/caption]
In particular we could use help from a Boston-area researcher, probably with pathology experience, who is enthusiastic about joining the PGP staff as a volunteer to create well-consented cancer cell lines. We get occasional emails from participants curious about donating surgical samples but we haven't had the bandwidth to take advantage of those opportunities. It's a string of tasks, mostly small but doable: organizing tissue sample kits and collections, shipping instructions, tissue dissections, cell line establishment, and getting the resulting cell lines to Coriell.
If you’re interested in joining us to create a new generation of well-consented cell lines, get in touch!
My apologies to the Personal Genome Project participants whose genomes are still "anonymous" and not yet added to their public profile. The CAGI team really wanted to provide competitors with one more month of time to make predictions, and so their challenge has extended to the end of DNA Day (11:59pm PST). We'll be able to publicly release these genomes to PGP profiles the day after that.
The Critical Assessment of Genome Interpretation (CAGI) challenge for predicting traits for 77 PGP genomes is still open for most of the month -- the challenge is scheduled to end on March 28. CAGI's latest news update also announces their conference will be in July, in Berlin, and lists several other challenges. There's still a lot of time to do analyses, visit their site if you're interested in participating.
Thanks again to the PGP participants who volunteered their genomes to be used by CAGI. Once the challenge is done your genomes will be linked from your public PGP profiles. We hope the PGP's uniquely public data facilitates dialogue and leads to publicly shared methods for genome interpretation.
A couple months ago I posted about our new trait surveys. Thank you to all the participants who completed these so far! I'm following up now with links to the data, a bit of Python code for interpreting them, and a little analysis.
The website has been updated to make the csv format files containing results from google surveys publicly available. Here are links to the PGP participant survey, and the twelve trait surveys:
To help anyone interested in parsing the data, I've shared the Python code I've used on Github. There's also a copy of the survey data as of Feb 23 there along with some demo code, and a Readme.
Finally, I did a bit of parsing of the trait survey data, combined with some features of the participant survey data (age and sex) to see if I could find anything interesting for you. The top 20 pairwise correlations below don't look terribly surprising, but I learned some new things. For example, I didn't know that TMJ disorder is much more common in women. (Of course, a quick web search discovers confirms this is a well known association.) This type of analysis isn't my forte -- maybe someone with more experience with machine learning can do cool stuff with this data!
(To read the table below: the first row indicates "60% of females reported a UTI, while only 12% of others reported one".)
| Trait 1 | Trait 2 | p-value1 | % 1 with 2 | % others with 2 |
|---|---|---|---|---|
| Female | Urinary tract infection (UTI) | 3.1e-23 | 60.3% | 12.0% |
| High cholesterol (hypercholesterolemia) | High triglycerides (hypertriglyceridemia) |
2.6e-15 | 36.9% | 3.1% |
| Female | Ovarian cysts | 1.9e-13 | 21.9% | 0.4%2 |
| 60+ years | Age-related cataract | 9.1e-13 | 36.8% | 2.8% |
| Female | Iron deficiency anemia | 2.4e-11 | 28.5% | 4.0% |
| Myopia (Nearsightedness) | Astigmatism | 5.7e-11 | 59.2% | 25.5% |
| High cholesterol (hypercholesterolemia) | Hypertension | 1.6e-09 | 42.9% | 11.6% |
| Iron deficiency anemia | Urinary tract infection (UTI) | 1.9e-09 | 69.2% | 25.3% |
| 60+ years | Age-related hearing loss | 6.5e-09 | 31.6% | 4.1% |
| Urinary tract infection (UTI) | Ovarian cysts | 1.7e-08 | 22.0% | 3.1% |
| Polycystic ovary syndrome (PCOS) | Ovarian cysts | 2.1e-08 | 66.7% | 6.6% |
| Hypothyroidism | Hashimoto's thyroiditis | 2.7e-08 | 23.1% | 0.6% |
| Temporomandibular joint (TMJ) disorder | Fibrocystic breast disease | 5.3e-08 | 28.6% | 2.4% |
| Nasal polyps | Chronic sinusitis | 7.0e-08 | 61.1% | 7.8% |
| Osteoarthritis | Bone spurs | 1.1e-07 | 29.8% | 3.6% |
| Female | Temporomandibular joint (TMJ) disorder | 1.4e-07 | 21.9% | 4.0% |
| Female | Fibrocystic breast disease | 1.9e-07 | 12.6% | 0.4%2 |
| Male | Hair loss (includes female and male pattern baldness) |
3.0e-07 | 29.5% | 8.3% |
| Carpal tunnel syndrome | Temporomandibular joint (TMJ) disorder | 4.4e-07 | 52.2% | 8.5% |
| Urinary tract infection (UTI) | Fibrocystic breast disease | 5.4e-07 | 14.4% | 1.2% |
1As calculated using a Fisher's Exact test. Note that these are not corrected for multiple hypothesis testing. I think a pessimistic Bonferroni correction would demand around 1e-6 for the magic 'p = 0.05' cutoff.
2I didn't look closely, but I suspect these non-zero numbers are because we have some transgender participants whose sex at birth differs from the gender they identify with (and the latter was what we have recorded on the participant survey).
 This is a guest blog post from Misha Angrist, Ph.D., an author, assistant professor at Duke University, and PGP-4.
This is a guest blog post from Misha Angrist, Ph.D., an author, assistant professor at Duke University, and PGP-4.
In the 7 February 2013 edition of Nature I have a commentary on genomic privacy arguing that it is time to re-frame how we think about this issue. I wrote this partly in response to the Science paper by Gymrek et al in which the authors used a combination of public genetic and genealogical data to re-identify the surnames of supposedly anonymous—or at least “de-identified”—people.
The initial reaction to my commentary from the senior author of the paper, Yaniv Erlich, was anger. He felt that I was denigrating his work, that I was implying that it had been done before and therefore it was not a big deal. Rereading my piece, I can see how he would think that; given a do-over and more space I reckon I would phrase some things differently. But I did not mean anything of the sort.
Yaniv and I had an honest and respectful exchange. His revised and measured response is here; I admire and appreciate his willingness to reconsider his initial reaction. I did my best to assure him that my goal was never to minimize what he and his team had done. On the contrary: Academic “privacy hackers” like Yaniv Erlich, Latanya Sweeney and Brad Malin are essential to understanding how secure our genomic data might—and might not—be.
What I was really trying to convey was frustration: we now have a decade’s worth of data demonstrating that genetic information is identifying. NIH (disclosure: I am a recipient of NIH funding), in its commentary that appears alongside the Erlich group’s paper, says it is concerned about this issue, but doesn’t seem all that willing to entertain policy alternatives that fundamentally challenge the status quo. The PGP, for example, is not mentioned.
Folks who study human beings are nervous. Indeed, in a phone conversation today Yaniv told me that a senior colleague of his said that his paper should not even be published lest it lead to a shutdown of public sequencing resources.
Really? So genomic identifiability is like Fight Club?
Three clarifications:
Since its founding, the Personal Genome Project has only accepted participants who understand and acknowledge re-identification as a potential risk. This "open consent" approach arose from our argument that privacy may be over-promised and that re-identification is increasingly possible as technology advances.
Dramatic progress in re-identification has been published today in Science (Gymrek et al.), and is reported on in Wired ("Scientists Discover How to Identify People From ‘Anonymous’ Genomes"). Wired's article features some quotes from George Church and highlights our project.
This is a guest blog post from James M. Turner, a Boston-area software engineer, freelance journalist, author, and PGP-65. James recently created a forum and wiki for discussion of topics related to the Personal Genome Project. While our staff isn't responsible for these sites, we plan to contribute to them and hope they provide an additional place for PGP participants to find useful information and answers to their questions.
When I was 8, I read "The Andromeda Strain", by Michael Crichton (yes, the geek force was strong in this one, even at a young age.) The book left a strong impression on me about the future of genetics, to the degree that I was writing programs in high school to convert DNA sequences to amino acids. Mind you, this was in the late 1970s, when you had to walk uphill both ways to school to save your BASIC programs on paper tape, but tell that to the kids today, I tell you...
For a while, I was sure that my future would be in the biosciences, perhaps as a geneticist. Unfortunately (or perhaps fortunately) for the biosciences, I was distracted from base pairs by the other up and coming technology of the time, computers. Although I continued to avidly follow the life sciences, I fell in love with software and have spent the last 35 years making hardware jump through hoops with clever code.
But a funny thing happened on the way to 2013. Genomics, and the obscenely steep slope of the $/genome price slide, has created another example of what trendy geeks like to call 'Big Data.' Big data is a challenge, because it strains the computational and storage limits of computers to analyze, but it's also an opportunity to correlate and draw new insights from datasets that used to live in their own private silos. The PGP, microbiome atlases, health data, exercise records, phenotypic traits, diet and much more are now digital and starting to hang around in the same neighborhoods. Between the years of 8 and 50, my two passions became "two great tastes, that taste great together."
I first became aware of the new field of personal genomics when I researched 23andMe and deCODEme for an article I wrote in 2009. As part of the research for the piece, I got my Single Nucleotide Polymorphism (SNP) data from 23andMe, while my wife tried out deCODEme. As anyone who has used the SNP services knows, there's interesting data to be looked at, but it's just a tiny fraction of what goes on in the entire genome. I had experienced a taste of my genome, now I wanted the real deal.
That's what led me to the PGP, drooling into test tubes, hanging out at GET 2012, and finally receiving my whole genome sequence a few months ago. Once I had all those lovely base pairs to play with, it immediately became clear that there's not a really good user manual for the data, a "Genome Interpretation for Dummies." I'm a pretty tech-savvy guy, and know enough about biology to be dangerous, but I quickly found myself dealing with the subtleties of GFF vs GTF vs BED format, comparison shopping genome browsers, and coming to the realization that a "whole" genome has small holes scattered throughout it (this must be why they call it shotgun sequencing...)
One of the things I know well from software is that crowdsourcing works. That's the entire model behind GET-Evidence, many eyes and fingers building up a larger and more useful database of gene to phenotype relationships, so that eventually a newcomer will have a wealth of information about their genome. But what's missing right now is a place to talk about the process, learn from each other, and share what works and what doesn't.
I started talking to the folks at the PGP a few months ago about the idea of setting up a forum and Wiki for PGP participants (and researchers, and anyone else who wants to join in) to share information, look for help, or just chat. For a number of reasons, it was decided that it would be better to have them hosted and administered outside of the formal PGP organization, and I was asked if I was interested in setting them up. I was, and have.
At http://forum.personal-genome.org/, you'll find the Personal Genome Project Forum. It's a place to introduce yourself, discuss the PGP, GET-Evidence, genomics, and anything else that you want to. The PGP is a community as much as a project, and people in a community should have a town square to mill around and chat.
For more formalized knowledge transfer, there's also http://wiki.personal-genome.org/, the PGP Wiki. Hopefully, this will grow into a fount of information about the how and why of genomes. I expect there will be a fair amount of cross-pollination between the Wiki and the forums, with forum discussions turning into Wiki articles, and people discussing wiki topics on the forums. There aren't a lot of rules at the moment (beyond the obvious about spam and privacy and civility), so the personality of the fraternal sites will evolve as people use them.
So, they're there, they're open for business, and with this blog posting, they're announced. A good first start would be to drop by the forum, register as a user, and put up an introduction in the appropriate topic. There's only two posts there right now, and it's a little lonely. We could also really use some articles on the basics of genomic data in the Wiki, I'm going to try to contribute as I have time, and some of the PGP staff have indicated there's stuff they'd like to write, but the more the merrier!
[caption id="attachment_964" align="alignright" width="240"] Canadian flag. Thanks to flickr user: ianalexandermartin (CC-BY-NC)[/caption]
Canadian flag. Thanks to flickr user: ianalexandermartin (CC-BY-NC)[/caption]
After several years of work, I am very happy to say that PGP-Canada has officially launched! I have had the great pleasure of working with the team led by Stephen Scherer at University of Toronto and the Hospital for Sick Kids to help organize a Canadian Personal Genome Project (PGP-Canada). This story stretches all the way back to July 2006 when George Church and his wife Ting Wu went to Toronto to speak about the Personal Genome Project. Read the press release from University of Toronto.
The Toronto Globe and Mail has created an amazing series about PGP-Canada and personal genomics generally called "Our Time to Lead: The DNA Dilemma": http://www.theglobeandmail.com/news/national/time-to-lead/
The content is really impressive in its scope, detail, balance and emotion. Especially touching are the videos exploring the human condition through the lens of individuals coping with disease and genetic risk. Altogether the series includes articles, videos, a poll, a digital game and other interactive material. A live debate about the risks and rewards of genetics research is schedule for December 18th.
For ease of navigating, I organized the content for you into 4 sets of links:
Series homepage:
Personal Genome Project and Public Genomics
Case Studies: Genetics in the Real World
These stories are touching and inspiring. I have to say that I'm compelled to give hugs to all of these people.
Background Information:
[caption id="attachment_952" align="alignleft" width="250"] Clifford Andrew (PGP-84) hiking on the Appalachian Trail in North Carolina (Photo posted with permission)[/caption]
Clifford Andrew (PGP-84) hiking on the Appalachian Trail in North Carolina (Photo posted with permission)[/caption]
This is a guest blog post from Clifford G. Andrew, M.D., Ph.D., Duke University School of Medicine, Class of 1972. Adjunct Assistant Professor of Neurology, Johns Hopkins University and PGP-84.
As a practicing physician, health protagonist and amateur genealogist, I have been an enthusiastic participant in the Personal Genome Project since enrolling in October 2010. This past fall, I returned to my alma mater, Duke University, for my Medical School Class of 1972 40th reunion. As in the past, we had set up a "mini-symposium" for discussion of various medical topics of interest to lay persons including spouses and families of our graduating class.
I decided that it would be appropriate to give a presentation on PGP to our group. As it turned out, our reunion was taking place within weeks after Robert Lefkowitz was announced as the 2012 Nobel Laureate in Chemistry for his research while we were at Duke on "G-Protein-Coupled Receptors." (1) In effort to put my talk in some context, I went back and did some research on previous Nobel laureates, and found to my surprise something which tied directly into the Personal Genome Project.
It turns out that 50 years earlier, the Nobel Prize in Physiology and Medicine for 1962 (while our group was still in high school) was awarded jointly to Francis Crick, James Watson, Maurice Wilkins "for their discoveries concerning the molecular structure of nucleic acids and its significance for information transfer in living material." (2)
In his Stockholm banquet acceptance speech, Watson was quoted as having said, "With our discovery of the structure of DNA, we knew that a new world had been opened and an old world, which seemed rather mystical was gone. At that time some biologists were not very sympathetic with us because we wanted to solve a biological truth by physical means. But fortunately some understood that by using the techniques of physics and chemistry, a real contribution to biology could be made."(3)
I went on to describe the Human Genome Project, first begun in 1990 with working draft in 2000 and final published draft in 2003, mapping for the first time the entire haploid reference genome for Homo sapiens containing 3 billion base pairs and 25,000 identified genes (but accounting for only 1% of the base pairs). (4) In 2007 James Watson was the second person to publish his fully sequenced genome online, stating that he did this "to encourage the development of an era of personalized medicine in which information contained in our genomes can be used to identify and prevent disease and to create individualized medical therapies." (5)
Indeed, that is what the Personal Genome Project is all about: using an innovative method for "open consent" in human subjects research, wherein all of the genomic data, as well as personalized details about environmental factors, and traits is posted on the internet, achieving a critical mass (105 individuals) of associated data, and allowing open access to physicians, medical scientists, and researchers for advancement of knowledge on how DNA sequence combine with environmental factors to result in clinical factors, traits, health and disease. (6)
In 1997, I was contacted by a Harvard researcher to participate in a 15 year study with 15,000 other physicians wherein we took four pills daily: vitamin C, vitamin E, beta-carotene, and multivitamin; or placebo for each. We reported annually on environmental traits, medications, and diseases. When the code was broken for the first three, I learned that I had been on PLACEBO for all, and that none of them influenced to any significant degree the incidence of cardiovascular disease or cancer. The study was concluded the month of the Duke mini-symposium, and I learned not only that I had been on the REAL multivitamin, but also that this had resulted in a small but significant 12% decrease in the incidence of overall cancer. (7)
The Physicians Health Study II was a demonstration of how evidenced-medicine is supposed to work. As meaningful as PHSII was, I expect the PGP to contribute several orders of magnitude more in terms of significant and long-term advancement of our understanding of how genetics and environment contribute to physical characteristics and health.
At Duke I passed out copies of the PGP pamphlet (8) and encouraged participants to enroll in the project. I am now doing the same with select patients of mine. I would suggest that each of us reach out to family, and friends to raise our numbers and get PGP to the 100,000 participant goal. Can you imagine what the GET Conference will look like in 10 years with numbers like that?
--
REFERENCES:
(1) http://www.nobelprize.org/nobel_prizes/chemistry/laureates/2012/
(2) http://www.nobelprize.org/nobel_prizes/medicine/laureates/1962/
(3) http://www.nobelprize.org/nobel_prizes/medicine/laureates/1962/watson-speech.html
(4) http://www.ornl.gov/sci/techresources/Human_Genome/project/about.shtml
(5) http://www.cshl.edu/Archive/watson-genotype-viewer-now-on-line
(6) http://www.personalgenomes.org/
(7) http://jama.jamanetwork.com/article.aspx?articleid=1380451
(8) http://www.personalgenomes.org/newsletter/pgp_flyer_longedgeflip.pdf
--
You may download a copy of the presentation here (1.2 MB PDF):
[caption id="attachment_935" align="alignleft" width="215"] The American Gut Project. Copyright Human Food Project, with permission.[/caption]
The American Gut Project. Copyright Human Food Project, with permission.[/caption]
Jeff Leach is an anthropologist by background – now an eager, albeit gray-haired graduate student at the London School of Hygiene & Tropical Medicine – and the Founder of the Human Food Project. In collaboration with Rob Knight at the University of Colorado-Boulder and Jack Gilbert at Argonne National Laboratory they have launched a large-scale citizen science project to document the diversity of the American gut microbial ecosystem – giving anyone in the U.S. the opportunity to participate and to compare their microbes with thousands of others with the hope of revealing patterns in diet and lifestyle that shape our microbial communities.
PersonalGenomes.org has partnered with American Gut to create a third party research opportunity for PGP volunteers. We recently sat down with Jeff for a brief Q & A:
Where did the idea for American Gut come from and why use crowd sourcing?
The idea was hatched out of some pilot research we were doing among traditional groups in Southern Africa. In 2011, we started laying the groundwork to characterize the gut microbiome of hunter-gatherers and subsistence farmers living on the edge of urbanization with the hopes of getting a glimpse at what our ancestral microbiome may have looked like before it ran gut first into the buzz saw of globalization. In other words, if we ever want to understand what a ‘normal’ or ‘healthy’ gut microbiome looks like – and how we might achieve that ancestral gut, again – we will need to look outside our modern world. So the idea was to assemble a large and diverse dataset of ‘modern’ microbiomes with extensive metadata (the American Gut Project) to get a handle on the diets and lifestyles that were potentially driving variability, to compare with more ‘untainted’ microbiomes around the world. Jeff Gordon, Maria Dominguez-Bello and others are doing important work in this regard as well.
Crowd sourcing was a great platform to launch this effort – quickly and efficiently. Citizen science has a long history and the gut microbiome lends itself well to public participation. As I believe food and health is this generation’s civil rights movement, social and crowd-like platforms for building a community, awareness and actionable solutions will emerge as great tools.
I see that a pretty impressive team has been assembled for the project
Yeah. The credit for that all goes to Rob Knight. He and Jack, along with others, launched the Earth Microbiome Project to analyze the microbial diversity of the entire earth. It’s the big-idea-can-do thinking that makes the American Gut project. The list of collaborators that have signed on to the project will participate via inter-lab/university agreements in the data interpretation. Some labs may also do some select portions of the analysis as well. The depth of the ‘collaborator bench’ will allow for some pretty interesting insight into the data as it comes available.
How can the PGP volunteers contribute or benefit from American Gut
No doubt your genetic makeup plays a significant role in the shaping your second genome (your microbiome). I think Ruth Ley from Cornell, who is also a collaborator on this project, is doing some interesting research in that regard. The PGP community is obviously very interested in their health and well-being and participating in the process. The detailed metadata collected with American Gut, coupled with the existing data from PGP, will make for some interesting insight. And the fact that PGP volunteers that join the American Gut project will be able to ‘claim’ their results, thus sharing of computable datasets between the two projects. This will be unprecedented.
We are very excited about this effort – but there is a window of opportunity that closes on January 7. So anyone that think they might want to help with American Gut, will need to do so quickly. Note also, first-in-first-out rule applies – ie, those who sign up the earliest will receive the quickest results.
Learn more and sign-up at www.indiegogo.com/americangut
12/1: There is a problem with the indiegogo site. They have been notified so hopefully it will be fixed soon.
12/3: Fixed! The American Gut page is live again after being down for the weekend due to some sort of administrative hiccup that required IndieGogo staff to intervene and nobody was available Saturday or Sunday. I guess I'm happy that IndieGogo staff got to take off for the weekend! :)
As Alex blogged earlier, the Personal Genome Project (PGP) is hoping to work with the National Institutes of Science and Technology (NIST) to use PGP materials (cell lines and DNA) for NIST's "Genome in a Bottle" reference material. One of the things NIST is looking for, and that we'd love to see more of, is diversity.
Seeking diversity
Because the PGP is self-recruiting, we don't have a very balanced set of participants. "Self-recruitment" means that all participants have enrolled in our project through word of mouth, finding our website and enrolling online. To put it bluntly, that means we mostly end up with young white men. Here are some graphs from our recent paper:
Researchers would love to see more diversity in PGP data. However, the self-recruitment model is ideally suited for the PGP: self-recruited participants are more likely to have a good understanding of the goals and risks involved. And so we’ll simply put out the word here for people already following us: underrepresented groups are especially appreciated. Research within one or two racial/ethnic categories isn’t necessarily a virtue, biracial and multiracial heritage may be even more interesting to some researchers and can open more areas for future research.
Why NIST wants PGP material
What does it mean for NIST to be considering the PGP for genome reference material? Major advances have been occurring in DNA sequencing and personal genomics; it's a competitive and rapidly evolving market. Manufacturers of instruments need standard human genome material to use for calibration of their machines, and others would like to use it to compare different devices and create common quality metrics. For example, the Food and Drug Administration may use this material for certification of sequencing instruments. It is possible this reference material will become ubiquitous -- spread far and wide, in a variety of commercial devices, with little ability to protect or regulate the uses of it.
[caption id="attachment_894" align="aligncenter" width="600"]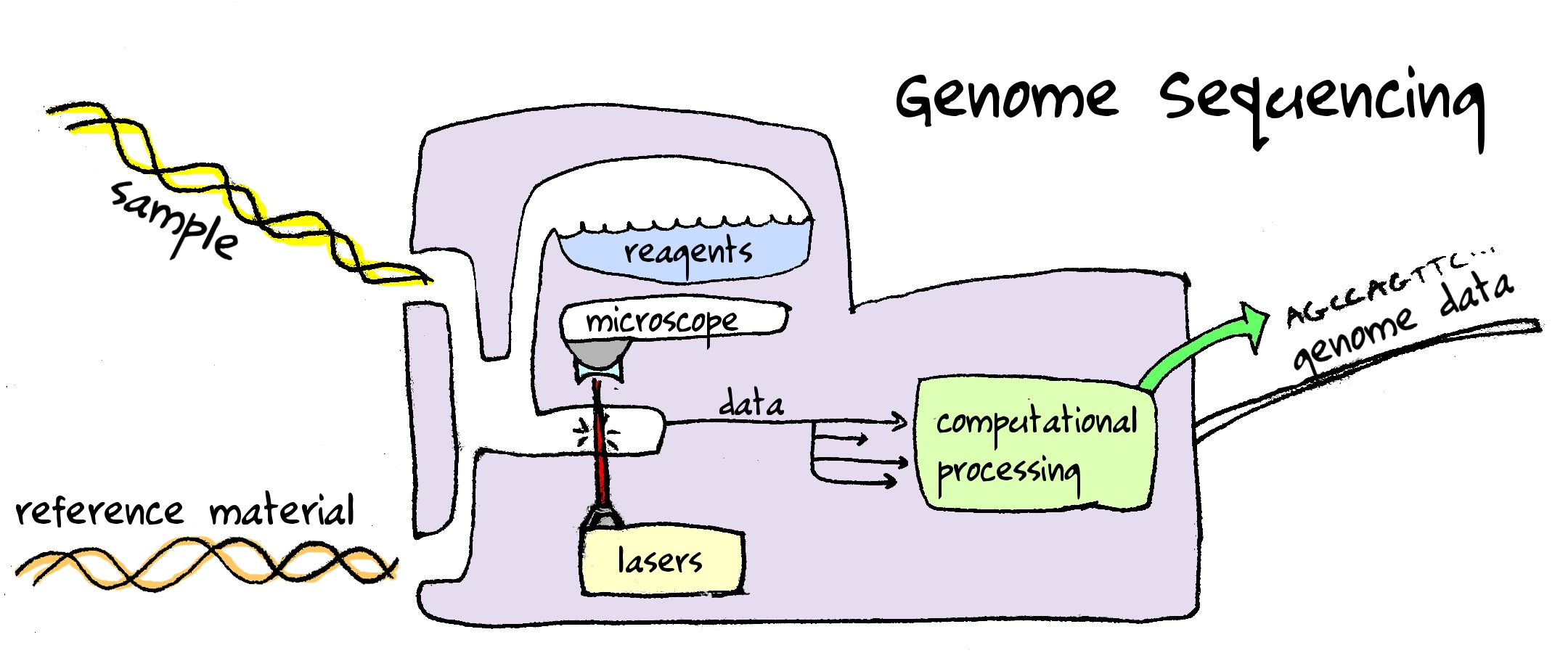 To visualize the potential widespread usage of reference material, I've made this informal sketch of the sequencing process. Not to scale, lasers may not be included in all models.[/caption]
To visualize the potential widespread usage of reference material, I've made this informal sketch of the sequencing process. Not to scale, lasers may not be included in all models.[/caption]
Why use PGP samples? Even though NIST's genome reference material will be manufactured using cell lines, those cell lines originally come from a person -- society is realizing that tissues and DNA are very personal things! In the wake of the experiences of Henrietta Lacks and HeLa cells (as documented in Rebecca Skloot's recent book), NIST wants to make sure the material they use comes from people who understood and agreed to potentially widespread usage. The PGP's "open consent" is a gold standard for careful consent to broad usage: PGP participants acknowledge and agree to things other subjects have not, including the risk of re-identification and commercial uses of their material.1
Parents and children
In particular, NIST is looking for "trios": two parents and a child. Researchers like to use samples from trios because they know every piece of DNA in the child comes from one of the parents. This makes it easier to assess error rates -- and that sort of quality control is what NIST expects the genome material to be used for. We think all such family groups are valuable, but current trios in the PGP haven't been the most diverse...
| Trio | Self-reported race/ethnicity |
|---|---|
| hu5D9DE3/huFE1569/huA8BCB0 | White |
| hu91BD69/hu38168C/huCA017E | Asian |
| hu16360E/hu28DA07/hu1A7894 | White |
| huAA53E0/hu8E87A9/hu6E4515 | White |
| huB4E01A/hu39790F/hu781C4E | White |
| huCDC3B8/huFE01E1/hu1E8957/hu961968 | White |
| huAA8CF9/hu7DB29E/hu2ED134 | White |
| hu620F18/huD4BF17/huD62596 | White, American Indian / Alaskan Native2 |
| hu1053CC/huFAF1FE/hu40D515 | Unknown |
| huC434ED/huD44B2B/hu25DE85 | White |
| hu36CDF1/hu210C97/huCFD87D | White |
NIST's team has told us they would like to have samples representing the breadth of human genetic diversity -- various ethnicities and multiracial heritages. Our project would love to enable that, but we are sensitive to the history of minorities in human subjects research. Participation in the PGP has many acknowledged risks and no promised benefits -- it definitely isn't for everyone. I can't even promise that NIST will use your samples (many would see that as a benefit rather than a risk). I'm simply going to write that NIST and other researchers wish they could have more diversity, and about the lack of it in the PGP -- maybe, if we're lucky, it will inspire some new participants to self-recruit.
Footnotes
1: More specifically, the PGP promises not to seek financial gain or commercial profit from materials (although cost recovery is allowed), but may "permit your cell lines to be used for research, patient care, commercial or other purposes". We don't expect anyone's genome to be uniquely valuable, but blocking any and all commercial uses of shared material is often viewed as overly restrictive. If a company wants to include NIST's DNA standard in a commercial machine, that would be a commercial usage of the material.
2: I have observed a high rate of people reporting both White and Native American ancestry in PGP participants (it's the second most common category, see the table below). While not questioning any specific individual, some genealogists have cast doubts on the high frequency of these cases. Elizabeth Warren's experience may be a common one.
| Self-reported race/ethnicity | # of PGP participants |
| White | 1285 |
| American Indian / Alaska Native, White |
40 |
| Asian | 38 |
| Hispanic or Latino, White | 24 |
| Hispanic or Latino | 21 |
| Black or African American | 12 |
| Black or African American | 12 |
| Asian, White | 11 |
| Black or African American, White | 7 |
| American Indian / Alaska Native, Black or African American, White | 4 |
| Hispanic or Latino, Black or African American, White | 3 |
| American Indian / Alaska Native, Hispanic or Latino, White | 3 |
[caption id="attachment_939" align="alignleft" width="294"] The Wildlife of Your Home. Copyright: Your Wildlife, with permission.[/caption]
The Wildlife of Your Home. Copyright: Your Wildlife, with permission.[/caption]
Rob Dunn is a biologist and writer in the Department of Biology at North Carolina State University. His lab group – in collaboration with Noah Fierer’s microbial ecology group at the University of Colorado–Boulder – have launched the Wild Life of Our Homes project, a continental-scale citizen science project that aims to build an atlas of house-associated microbial diversity. Dunn and his team see the homes of North America as the next ecological frontier. They aspire to understand how the physical characteristics of a home, its inhabitants, and the landscape in which it is situated influence the microbial communities that live there. Moreover, they will investigate the reverse: how the presence or absence of home microbes may influence our own health and well-being.
PersonalGenomes.org has partnered with Dunn and Wild Life of Our Homes to create a third party research opportunity for PGP volunteers. We recently sat down with him for a brief Q & A:
How did you get interested in the microbes of homes?
I guess this story has multiple answers. My first interest was in the context of writing. I was writing The Wild Life of Our Bodies and became really interested in what we do and don't know about the species we interact with every day. I was struck by how little was known about the species in our homes. No one has ever exhaustively surveyed the species of homes. No one even has a good list of the animals present, much less the smaller beasts. This fascinated me. What fascinated me even more than writing about this problem was that I could do something about it and so we began the Wild Life of Our Homes and other related projects in which we work with the public to study one of the least known but most important habitats on Earth, your home.
What is your big vision for Wild Life of Our Homes?
It depends on the day I guess, but when I am feeling ambitious I think we might be able to pull off the most complete survey of the insides of our homes ever achieved and do it in such a way to understand what determines the species that live with us. The big, big, vision is then to move from understanding who is present and why to being able to garden species that benefit us, whether they are animals, plants, fungi or bacteria. We are good at killing species we think are bad, far less effective at gardening species that benefit us (with the exception of our foods).
What special contribution do you think PGP volunteers could make to the Wild Life of Our Homes project?
Oh, well this is exciting. One of the really interesting things to think about in the context of microbes in homes is the interaction between our bodies and their cells. We know that many (but not the majority) of the species living in our homes depend on our bodies and their skin and other bits for food. And so presumably the species that live on you are influencing the species we find in homes. But the really interesting question is whether your genes are actually influencing the species on you and the species floating around you—on your pillow, on your cutting board, anywhere else. Is the composition of your house influenced by the extended influence of your genes? That seems very conceivable but is hard to test. With PGP data we will be able to perform the test. We can also consider the reverse. To what extent do the microbes in your house influence your health and well-being and how much is that effect contingent on your genes. The rich data provided by PGP volunteers will really be wonderful in the ways that it will allow us to think of human and home as part of a continuous ecosystem.
Why recruit citizen volunteers to accomplish your research goals?
It is literally the only way we can see what is going on. I used to study rain forests. Sometimes it was hard to get to a field site. There were narrow trails, dangerous snakes, malaria parasites, and sketchy buses, but you could get there. When it comes to studying bedrooms on the other hand, access is more difficult. More to the point, even if we could go into 1000 bedrooms, we can go once. The folks we work with can study their homes every day. That, to us, is the great thing, to be able to form a network of public scientists each of whose houses becomes a kind of long-term ecological research site.
I understand you’re in the process of analyzing data from a pilot study of the microbes living on surfaces in 40 homes in North Carolina. What have you learned so far? Have there been any surprises?
Microbiologically, we can't tell toilet seats and pillowcases apart. I don't know how that changes my life, but it is true. Also, there are strong and discrete habitats within the home microbiologically speaking, but the big surprise is that there are big differences among houses (just as we have seen in another study, among belly buttons). The fun question, the one we are enlisting PGP participants to help us with, is explaining what accounts for those differences. Outdoor climate? Backyard biodiversity? Your genes? Your dog? Your carpet? The type of house you live in? Any and all of these things might matter, but our data so far suggest that most of them don't. Our preliminary data do suggest that some aspects of the ecology of our homes may be simpler than we anticipated, but we need to see more houses. We need to test our anecdotes against what we see across North America from sea to shining sea, or should that be from toilet seat to shining toilet seat?
Wild Life of Our Homes is just one of many public science projects spearheaded by Dunn and his team. You can learn more about their work at http://www.yourwildlife.org/
Important Note for PGP participants
PGP volunteers interested in participating in the Wild Life of Our Homes project should log into their PGP account and visit the third party page to register. We strongly encourage PGP volunteers to sign-up for the project in this way so that you may easily link your home microbiome data to your public PGP profile once the data is available.
Part of what makes Personal Genome Project participant data uniquely valuable is our publicly shared trait data connected to public genetic data. A year ago our project was frustrated when our best resource for importing health data -- Google Health -- was discontinued. Ward soon got an interface running to import data from Microsoft HealthVault; the CCR-format data it produces is very similar, but isn't trivially combined with our Google Health records.1 We wanted to improve the quality of our trait data and provide another option for adding traits to public profiles.
And so we created a set of twelve trait surveys (the links below will only work for PGP participants) covering 239 traits and diseases:
To select what traits and conditions to include, the Google Health data was an invaluable resource. I was able to combine conditions using their ICD-9 codes (or, if unavailable, by internal Google codes).2 Here's the five most common reported traits:
[caption id="attachment_827" align="aligncenter" width="540"]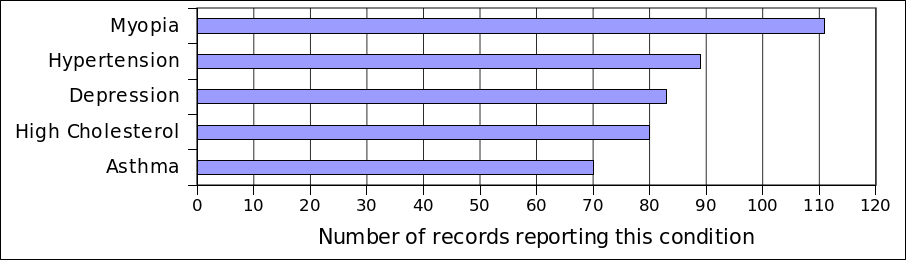 Top five conditions reported on Google Health records contributed by PGP participants.[/caption]
Top five conditions reported on Google Health records contributed by PGP participants.[/caption]
We tried to settle on four encodings corresponding to each trait: ICD-9, ICD-10, SNOMED CT, and NCIMetathesaurus CUI.3 I've shared our list of traits surveyed, along with the encodings we consider them associated with, as a Google spreadsheet.
A useful aspect of the ICD encodings is their organization by topic, and so our traits were split into twelve survey topics by ICD-9 encoding. It's impossible to be perfect in a first pass, but we tried to include anything that was fairly well-defined, not too rare (a prevalence of at least 1 in 10,000), and within the twelve ICD-9 ranges selected for the surveys. You might notice that some ICD-9 ranges were not used -- most notably, the category of mental traits and disorders. We do hope to survey these as well, but I want to be sure that participants are able to easily manipulate data on their public profiles before adding such a potentially sensitive category.
All PGP participants are invited to enter public trait data using these surveys -- although contributing such information is optional, and not required for participation. Even if you don't see a condition listed in the survey that you want to add, submitting an empty survey is useful information. I hope to follow up soon with a blog post analyzing some of the resulting data.
1On top of it, these records contain identifying data (like names and email addresses) that our participants weren't intending to make public. This meant we couldn't share the raw data, anything we shared was limited by our private CCR data interpretation process. Ideally we wouldn't be in this position: sharing raw data and allowing others to interpret it would be better, scientifically.
2If you're interested in it, this data was made available as "Dataset S1" in our recent open-access PNAS publication.
3Why four different coding systems? A couple of reasons: for redundancy, to facilitate using our data in other systems, to provide a starting point for harmonizing data from imported health records, and because we weren't (and still aren't) sure whether or how we'll be able to work with the licensing issues associated with some of these popular encoding systems.
I've uploaded a video of the talk I gave at the 2012 Open Science Summit. The talk itself is only twelve minutes long -- it's a fairly fast-paced overview of the Personal Genome Project's motivations and goals, with updates on recent progress.
[youtube http://www.youtube.com/watch?v=Z0aoYMdoZ04]
DNA donated by Personal Genome Project participants may be chosen by the National Institute of Standards and Technology (NIST) to become reference materials for new human genome sequencing standards!
NIST's "Genome in a Bottle" consortium convened in August to initiate the establishment of a human genome standard. This "meter stick of the genome" will serve as an international reference for identifying variation across individual genomes, and be used to establish professional standards for clinical human genome sequencing. Specimens donated by PGP volunteers are viewed as ideal candidates to serve as these new reference standards due to the depth and availability of public PGP datasets as well as the strength of the consent process used in the Harvard PGP study.
Nothing has been finalized yet, but this may become an exciting opportunity for our participants to contribute to an effort to standardize a new and rapidly-evolving field of genomics and personalized medicine. The program is specifically interested in the participation of parent-child trios (including both parents and one or more offspring).
I'm going to be flying to Mountain View this evening to attend the Open Science Summit. Various PGP members have attended and spoken in previous years; this year I'm up and scheduled to give a 15-20 minute talk Saturday morning (in the 10:45am-12:00pm PST grouping).
Open science is of course one of our core motivations as a project, and I look forward to meeting many like-minded folks there. Live streaming of the summit should be available at this site: http://fora.tv/conference/open_science_summit_2012/livestream
For twitter users, the hashtag du jour will be "#OSS12". Last minute tickets if you want to attend in person are here: http://opensciencesummit2012.eventbrite.com/ (looks like $300 at this point).
The Personal Genome Project is working with the Critical Assessment of Genome Interpretation (CAGI) this year to provide a genomic interpretation challenge using PGP data! CAGI's use of PGP data is a demonstration of how publicly sharing genome & trait data is invaluable to science: because the data is public, the challenge is open to everyone. No restrictions or requirements need to be met to access the data.
How will the challenge work? In the upcoming week or two we will be returning genomes to some participants. Currently genomes automatically become public after 30 days of private access, but participants have the ability to publish a genome immediately should they choose to do so. We've added an additional option for CAGI: participants can release the genome to be used as a CAGI genome. When they do this, the genome becomes public -- but which participant account the genome belongs to is kept secret. At the end of the challenge genome data will be linked to the specific participant account.
Of course the other half of CAGI is the predictions -- we need trait data from participants for the CAGI researchers to try to predict! I've been working hard the last couple weeks to make a set of trait surveys for PGP participants. These surveys aren't just for CAGI, they're for all participants and they'll remain open after CAGI is ended. For a genome to be used by CAGI a participant will need to complete all the surveys, but all participants are encouraged to fill them out.
The trait surveys are publicly shared and entirely optional -- only choose the items in the survey that you want added to your public profile. You can find the surveys ("PGP Trait & Disease Survey 2012") at the bottom-center of the screen when you log in to your PGP account.
I am a PGP director and a participant, and in the latter role I received a recent email that urged me to “READ THIS!” It came from a family member and was triggered by the second segment in a recent series on personal genomics featured on National Public Radio’s Morning Edition (in a previous post Madeleine Ball highlighted the first segment featuring the PGP and George Church). The email-triggering segment featured two scientists whose genomes have been sequenced: James Watson, co-discoverer of the structure of DNA, and Mike Snyder, Director of the Center of Genomics and Personalized Medicine at Stanford University. Both Watson and Snyder presented generally supportive views of the process, but Snyder’s story provides more important lessons about the ups and downs of biomedical self discovery.
Beginning with the upside of Professor Snyder’s story, the publication featuring his genome sequence and other “omic” data provides strong evidence that at least some genomic predictions are both accurate and actionable (1). He learned he has an elevated risk of basal cell carcinoma, hypertriglyceridemia, and Type 2 diabetes (T2D). Upon learning of these risks he began to be monitored for these conditions. Consistent with the genomic prediction he did have high triglycerides, and the problem was successfully medicated.
The most interesting biomedical subtext of Snyder’s story began about a year after the prediction of elevated risk of T2D (due to variants in 3 genes). During the first year his blood glucose and proportion of glycated hemoglobin (HbA1c) were normal (HbA1c is a glycated form of hemoglobin, a protein in red blood cells that aids in oxygen transport; glycation is non-enzymatic attachment of glucose to the protein, which is a measure of persistently high blood glucose levels). Immediately following a viral infection, his blood glucose rose rapidly and in less than a month he had full-blown T2D as measured by high blood glucose (above 126 mg/dL), and about a month later as measured by HbA1c. His levels remained high for about two months after which he changed his diet and began to exercise more, and after six months his glucose and HbA1c levels returned back into the normal range. Overall, his fasting glucose level exceeded the threshold value for a clinical diabetes diagnosis for about 4 months.
But the story doesn’t end there, and this is why I received the exclamatory email. Even though Professor Snyder’s triglycerides and glucose are under control, insurance complications arose from the initial T2D diagnosis. According to NPR:
“After sequencing revealed his high risk for diabetes, his wife tried to increase his life insurance. But because of that high risk, the price shot through the roof. ‘So the bottom line is my life insurance ... essentially became prohibitively expensive,’ Snyder says. Federal law bans health insurance companies and employers from penalizing people based on genetic information, but the law doesn't apply to life insurance or long-term care insurance—leaving people like Snyder vulnerable to discrimination.”
Since there was some uncertainty about this NPR report, I asked Professor Snyder for his input and he emailed me the following clarifications:
The NPR story is correct that GINA (the Genetic Information Nondiscrimination Act of 2008) does not protect consumers against genetic discrimination when they purchase life or long-term care insurance. However, the story appears to mischaracterize the causal chain of events in this case: Snyder’s prohibitively high life insurance rate did not result from the discovery of risk alleles in his genome; it resulted from an old-fashioned medical diagnosis of diabetes by his physician based on clinical tests that showed high levels of diabetes-specific biomarkers (2). And despite his insurance problem, Professor Snyder believes genomic self discovery is more positive than negative and that his genome sequence helped him deal with his diabetes in a timely fashion.
It is fairly obvious why Snyder and his family would be happy with the present biomedical outcome but here is something that isn’t quite as obvious: his frequent testing helped him to chart a data-guided course to recovery, but it also made detection of his transiently elevated glucose much more likely than, say, annual testing. A reasonable estimate is that it made it about twice as likely since he had elevated glucose for about four months and sub-threshold levels for the remaining 8 months of the year. So, in addition to potential benefits, there might be a downside to extremely frequent, elective testing.
Professor Snyder’s story reminds us that the builders and first travelers on the road to data-driven healthcare occasionally experience remarkable and previously unknown vistas of self determination, but they also face uncertainty and risk. Many risks can be avoided or managed, but science is often a murky business and news reports on these difficult topics can be misleading, further obscuring the way forward. Our loved ones’ anxieties are heightened by the occasional fog of confusion—triggering emails or discussions of concern, or even alarm. But now that we see the relevant facts in this case—and the red flag posted clearly over one hazard in the road—we also can see ahead more clearly as we push on toward the greater goals of this unprecedented collective experiment.
References and Footnotes
1) Personal omics profiling reveals dynamic molecular and medical phenotypes. Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HY, Chen R, Miriami E, Karczewski KJ, Hariharan M, Dewey FE, Cheng Y, Clark MJ, Im H, Habegger L, Balasubramanian S, O'Huallachain M, Dudley JT, Hillenmeyer S, Haraksingh R, Sharon D, Euskirchen G, Lacroute P, Bettinger K, Boyle AP, Kasowski M, Grubert F, Seki S, Garcia M, Whirl-Carrillo M, Gallardo M, Blasco MA, Greenberg PL, Snyder P, Klein TE, Altman RB, Butte AJ, Ashley EA, Gerstein M, Nadeau KC, Tang H, Snyder M. Cell. 2012 Mar 16;148(6):1293-307. PMID: 22424236
2) It is important for PGP participants to understand that PGP-generated data and reports are not equivalent to a clinical test and, according to the PGP consent form “are never intended to substitute in any way for professional medical advice, diagnosis or treatment. You may not use any PGP-generated report or any other PGP-supplied data or results for any medical or clinical purpose until you have confirmed the relevant sequence, data, interpretations and/or findings with a licensed healthcare professional.” Nevertheless, just because these data and reports are not clinical diagnoses doesn’t guarantee that they can’t or won’t be used to make decisions that might adversely affect you.
There was a mention of the Personal Genome Project in a story by Rob Stein on NPR yesterday: "As Genetic Sequencing Spreads, Excitement, Worries Grow". In the context of the vastly reduced cost of personal genomes, George Church discusses the need for publicly shared personal genome and health data.
In addition to the promise of personalized medicine, the story also highlights privacy concerns. These privacy concerns are reflected in how most other genome research projects are conducted: it is very difficult for most researchers to share data, because research subjects have been promised privacy and this needs to be protected. In contrast, PGP participants have agreed to the public sharing of their data -- an unusual waiver of privacy guarantees, made only after they demonstrate an understanding of the risks. This is creating an invaluable public resource, critically needed for scientific progress in this rapidly developing field.
Many thanks to the PGP participants who have already donated exome data! We are working to feature these and other donated data on our site.
Customers using 23andme's pilot exome service may have recently received an email from 23andme notifying them that online hosting of their exome data will end soon (at the end of August). Before this occurs, we would like to communicate to our participants how they can donate this data, if they wish to do so.
It is ideal to have the full data set donated (BAM data as well as the VCF file). These files are large and difficult to upload. We believe the easiest method for participants to donate this data is to provide the Personal Genome Project with the information needed for download (as given to you by 23andme):
(1) the Amazon Web Services URL
(looks like: https://exome-export.s3.amazonaws.com/AB1234_C56DE78a9b.tc)
(2) the decryption password
(looks like a random string, e.g. "XahN7tah4s")
With this information we can download the data directly and decrypt it. An email from you containing this information will be treated as a "public donation of data" to the PGP and may be made public immediately. (Making the BAM data public may take some time though. You can check the status of things by seeing what is visible on your public profile.)
To email us, please log in to your participant account on my.personalgenomes.org and click the "Contact Us" button. By receiving the information through your participant account, we can confirm that the donation was personally made by the participant.
Please let us know if you would rather share the data with us in some other manner. Thanks again to all our participants!
[youtube=http://www.youtube.com/watch?v=f3WF8-K4a2k]
Latanya Sweeney: MyDataCan.org (watch video) as presented at the 2012 Genomes Environments and Traits Conference at Harvard Medical School. Dr. Latanya Sweeney, Ph.D. is Visiting Professor and Scholar, Computer Science Director, Data Privacy Lab, Harvard University. See also Dr. Sweeney's recent testimony to the Presidential Commission for the Study of Bioethical Issues on the topic of genomics and privacy.
One of the most significant publications in the history of Alzheimer’s Disease (AD) research—and in genomics research generally—was just published online by Jonsson and colleagues in Nature magazine (1). I base this lofty assessment on impressive advances of two general types made by this study: those specific to AD, and others of even greater and more general importance. To whet your appetite for a more full taste of the latter, consider that the Nature title, “A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline,” is compelling, but greatly undersells the magnitude of the discovery within. The logically equivalent title of this blog post, on the other hand, highlights the underlying reason this discovery is so notable.
More about this shortly, but, for now, let’s back up and consider how this study contributes to our understanding of the biology of AD (if you're not really interested in AD, but want to understand the far-reaching consequences of this discovery, you can jump to the next section below). The study reveals the identity of a variant of amyloid precursor protein (APP) that is protective against both AD and general cognitive decline. This variant results from an amino acid substitution of a threonine (T) for alanine (A) at amino acid position 673 in the APP protein near a key processing site involved in the production of amyloid beta (the variant is known as APP A673T). Therefore, when considered together with previous studies, this study appears to validate beyond doubt the previously controversial amyloid beta cleavage and aggregation hypothesis.
This is the first such protective minor variant [f1] ever discovered, and it previously had gone undetected because it is so rare—present in less than 1% of Icelandic and Scandinavian populations, and even rarer in North Americans. Bear in mind that APP A673T might exert maximum protective effects on Icelanders (Icelandic genetics and environments are unusual; the potentially complex interactions of genes and environments highlights the importance of the broadly inclusive information collection and aggregation approach taken by the PGP (2)). Nevertheless, for reasons explained below, we should expect either validation of the protective effects of this genetic variant or the discovery of other similar ones in non-Icelandic populations.
For context, contrast APP with APOE, the best known gene associated with increased risk of AD. APOE e3, the primary or major variant of APOE, is protective against AD, while the minor variant APOE e4 increases risk. The average worldwide frequencies of e3 and e4 are about 78% and 15%, respectively (3). This is considered to be the more typical case of a minor variant increasing risk of disease [f2], and deleterious minor variants are the focus of most recent studies and gene hunters. Also in contrast to APOE, APP variations from the typical alanine at amino acid position 673 (673A) appear to be extremely rare among humans; the results of Jonsson and colleagues suggest that the frequency of this predominant variant might represent about 99% of all variants at this amino acid position.
About 2% of the population are APOE e4 homozygotes and they are reported to have a 10 to 30-fold increased risk of AD relative to people who carry no e4 variants , so discovering this about yourself or a loved one can be quite worrisome (f3). However, Jonsson and colleagues report that APP A673T protection against AD appears to extend even to those with APOE e4 genotypes. The protective effect of APP A673T appears dramatic in other ways, too. The number of carriers who have undergone routine cognitive testing is small (and therefore current analyses are subject to great uncertainty), but one analysis of cognitive decline performed in this study indicates at least a 10 year cognitive advantage of carriers versus non-carriers between ages 80 and 100 years old.
Large-scale genome sequencing is shifting the discovery paradigm
It is important to consider the approach used to make the initial finding in this groundbreaking discovery: whole genome sequencing of a large number (1,795) of people. The belief that whole genome sequencing has a good chance of succeeding where other approaches [f4] have failed has met with increasing recent skepticism by some scientists (4). The discovery of APP A673T is a clear vindication of genome sequencing as a tool for discovery of both potential causes and cures of disease and debility [f5], and it underscores the existence of an often unrecognized but extremely important class of genetic variation: functional minor variants that provide protection against common diseases.
Some of these variants are likely to be so rare that large-scale genome sequencing is the only efficient means of discovering them. As I mentioned above, APP variations at alanine 673 appear to be quite rare in human populations—and this rarity probably extends far beyond humans. I performed a quick analysis of the evolutionary conservation of this alanine across many species and found that it is conserved across all mammals, including dolphins. It also appears to be present in chickens, turkeys, anolis lizards, turtles, and at least some zebrafish, although it is not found in many other fish. Given this level of evolutionary conservation we can infer that amino acid substitutions at this position are strongly selected against in mammals and certain other organisms. It is very interesting and might be a bit surprising that a rare minor variant has such a strong protective effect relative to the predominant variant—and it is also why this discovery is so important.
Returning to the title of this post, consider the flipside of this protective phenomenon: the highly conserved and most common variant of APP increases the risk of AD and cognitive decline. But how is this possible?
How can extremely common variants be deleterious?
It is commonly assumed that natural selection purges most deleterious variants from the overall gene pool. This has been mostly true throughout human history, but nowadays most people live to ages far in excess of those shaped by evolutionary forces over that time. Gerontology is the study of aging and senescence, and gerontologists have accepted for decades that natural selection exerts diminishing selective pressure on the genes of an organism transitioning from reproductive to post-reproductive age.
In 1952 Peter Medawar proposed that deleterious variants will arise and persist in a population if negative effects occur very late in life (5). In 1957 George C. Williams extended this general framework with a critical insight: variants that contribute essentially to high levels of reproductive fitness early in life, but that also carry late-life deleterious effects, will persist through positive selection—even if these later effects are catastrophic (6) (Williams’ theory is often called Antagonistic Pleiotropy). In summary, there are two general and non-exclusive ways that gene variants (even very common ones) might exert negative effects in the post-reproductive period of life: 1) the absence of selection against certain variants because their late-life negative effects have little or no impact on reproduction, and 2) positive selection for variants that contribute reproductive benefits early in life with the unfortunate side effect of negative consequences after reproduction has ceased.
Common diseases and common variants reconsidered
When properly framed in this way, it is easy to understand how the vast majority of people might carry variants of APP that predispose to AD and cognitive decline late in life, while a rare minor variant is protective. We don’t yet know whether or how the more common APP variant is beneficial earlier in life relative to the A673T variant, but it is clear that A673T carriers are overrepresented in later life by several fold relative to non-carriers. One possibility for the evolutionary conservation of alanine 673 is that it carries substantial early life and reproductive benefits, especially in historical or more natural environments. It will be interesting to compare the reproductive successes and other phenotypes of the people carrying different variants.
It is important to understand that the evolutionary framework constructed by Medawar, Williams, and others has wide-ranging implications for the genetic determinants of disease and debility in older people. Most people who develop serious disease in developed countries are older and a few related diseases lead to the majority of mortality (cardiovascular diseases and cancer together account for about half of all mortality in developed countries). For non-gerontologists this discovery of APP A673T by Jonsson and colleagues provides a new twist in the public scientific discussion on common diseases and common variants. We should expect that the protective effects provided to older people are unlikely to be limited to this variant or to Alzheimer’s disease, and as we sequence more genomes we’ll surely discover more such variants protective against other diseases and general debility. Each time we do, there will be a logical complement or corollary for common and even predominant variants: they are risk factors for disease.
Protective variants provide a direct route to therapies
the most important information any protective variant provides is to reveal a specific way biology mitigates an extremely serious health risk, and therapies can be designed to reproduce its critical function. Pathogenic variants aren’t nearly so helpful. In the case of APP, drugs that mimic the effect of A673T have been in development for several years already (7) but no such drug is yet on the market or even in late stage clinical trials, but promising candidates are in early trials. The reason these drugs are already in the pipeline is that decades of basic biology had already pointed to one promising way to treat AD; so whole genome sequencing isn’t solely responsible for the eventual development of these therapies. However, given the very large protective effect of A673T for both AD and general cognitive decline, the market for mimetic therapies might be much larger than previously estimated—and their individual and societal benefits would be profound.
FOOTNOTES:
[f1] A major variant is relatively common in a population and a minor variant is uncommon.
[f2] Typical examples of protective minor variants provide protection against pathogen infection, often through loss of function (see Madeleine Price Ball's Invulnerability to Stomach Flu Is My Secret Superpower). APP A673T is a functional minor variant which has different effects on a carrier’s intrinsic biochemistry and physiology relative to the major variants, so should be considered differently than variants selected for pathogen resistance.
[f3] Some reports suggest that the influence of APOE e4 on AD might be largely mitigated by routine exercise (8)
[f4] For example, genome-wide association studies (GWAS) using single nucleotide polymorphism (SNP) data.
[f5] Debility means frailty, poor health, or weakness, especially resulting from old age.
REFERENCES:
1. Jonsson, T., Atwal, J.K., Steinberg, S., Snaedal, J., Jonsson, P.V., Bjornsson, S., Stefansson, H., Sulem, P., Gudbjartsson, D., Maloney, J. et al. (2012) A mutation in APP protects against Alzheimer's disease and age-related cognitive decline. Nature, Advance online publication.
2. Ball, M.P., Thakuria, J.V., Zaranek, A.W., Clegg, T., Rosenbaum, A.M., Wu, X., Angrist, M., Bhak, J., Bobe, J., Callow, M.J. et al. (2012) A public resource facilitating clinical use of genomes. Proceedings of the National Academy of Sciences, USA, Advance online publication.
3. Eisenberg, D.T., Kuzawa, C.W. and Hayes, M.G. (2010) Worldwide allele frequencies of the human apolipoprotein E gene: climate, local adaptations, and evolutionary history. American journal of physical anthropology, 143, 100-111.
4. Roberts, N.J., Vogelstein, J.T., Parmigiani, G., Kinzler, K.W., Vogelstein, B. and Velculescu, V.E. (2012) The predictive capacity of personal genome sequencing. Science translational medicine, 4, 133ra158.
5. Medawar, P.B. (1952) An unsolved problem in biology. HK Lewis and Co, London.
6. Williams, G.C. (1957) Pleiotropy, Natural Selection, and the Evolution of Senescence. Evolution, 11, 398-411.
7. Vassar, R., Kovacs, D.M., Yan, R. and Wong, P.C. (2009) The beta-secretase enzyme BACE in health and Alzheimer's disease: regulation, cell biology, function, and therapeutic potential. Journal of Neuroscience, 29, 12787-12794.
8. Head, D., Bugg, J.M., Goate, A.M., Fagan, A.M., Mintun, M.A., Benzinger, T., Holtzman, D.M. and Morris, J.C. (2012) Exercise Engagement as a Moderator of the Effects of APOE Genotype on Amyloid Deposition. Archives of neurology, Epub ahead of print.
[youtube=http://www.youtube.com/watch?v=fhEeztPLG0I]
Gregory Stock: Biomarkers for Mercury Toxicity (watch video) as presented at the 2012 Genomes Environments and Traits Conference at Harvard Medical School.
I’m proud to announce the open access publication of the first major research paper for the Personal Genome Project, published as an Inaugural Article in the Proceedings of the National Academy of Sciences: “A Public Resource Facilitating the Clinical Use of Genomes” by Ball, Thakuria, Zaranek, et al.
This paper ties together several related stories in the PGP’s research. It is an introduction to the PGP participant data as a public resource, it discusses some of our experiences with the pilot PGP-10 genomes, and it details our development of GET-Evidence in response to these experiences.
PGP participant data
Many reading this post already know it: the PGP has an exciting group of participants who have volunteered to make some highly personal data public for the sake of research. Our paper reviews the innovative open consent method that made this possible -- by publicly sharing our human subjects research protocols, we hope to encourage other projects to adopt similar methods for publicly shared genomes and other re-identifiable research data. We highlight the many participants who have demonstrated their commitment by publicly sharing data from electronic health records and describe the PGP-10 genome data we have released.
Experiences with the pilot PGP-10 genomes
As we release genome data publicly we are also returning that data to participants -- and, to provide participants with some understanding of what they are making public, we try to give an interpretation of their genome data. (Albeit a very rudimentary, tentative interpretation which almost certainly contains gaps and errors.) Thus, genome interpretation has become one of the core areas the PGP has focused on. The field is in its infancy though, and so far whole genome interpretation efforts by other groups have focused on discovering new disease-causing variants in people believed to have genetic disorders. Whole genomes have been effective for discovering new disease-causing variants, but most PGP participants (and most people!) aren’t believed to have serious undiscovered diseases. What happens when you interpret genomes of presumed-healthy people?
What happened was that we found several rare variants predicting diseases our participants didn’t have. Sometimes these were scary! The variant MYL2-A13T in PGP6 (hu04FD18, Stephen Pinker) was predicted to cause hypertrophic cardiomyopathy. The variant SCN5A-G615E in PGP9 (hu034DB1, Rosalynn Gill) was predicted to cause long QT syndrome. Both of these are late onset diseases that the participant could be unaware of, and could cause sudden death.
Some variants predicting severe effects in the PGP-10
| Participant | Variant | Putative effect |
|---|---|---|
| PGP5 (hu9385BA) | PKD1-R4276W | Autosomal dominant polycystic kidney disease |
| PGP6 (hu04FD18) | MYL2-A13T | Hypertrophic cardiomyopathy |
| PGP9 (hu034DB1) | SCN5A-G615E | Long QT syndrome |
| PGP10 (hu604D39) | PKD2-S804N | Autosomal dominant polycystic kidney disease |
| PGP10 (hu604D39) | RHO-G51A | Autosomal dominant retinitis pigmentosa |
These predictions couldn't all be correct; the PGP-10 couldn’t possibly have all of these diseases. In the process of interpreting these genomes and reviewing genetic variants, we developed a system for reviewing variants that critically examines the evidence for the variant -- not merely how bad the putative effect is, but how strong the evidence is supporting that hypothesis.
GET-Evidence: a system for personal genome interpretation
To facilitate the process of genome interpretation, we have created the Genome-Environment-Trait Evidence (GET-Evidence) system. Genome analysis is facilitated by GET-Evidence in a two step process: variants are prioritized for review, and then the review of a variant is recorded and used to create a genome report.
Prioritizing variants for review combines two reasons that one might want to pay special attention to a variant: the existence of published information associating the variant with an effect, and a computational prediction that the variant is disruptive and more likely to cause disease. As a result, the system combines interpretation based on existing knowledge with the potential for discovery of new disease-causing variants.
Variant interpretation then occurs through variant pages which gather numerous resources assisting the review process: variant frequency, computational predictions, and links to external databases. An editor can then add information to the variant’s page, including: the variant’s effect, inheritance pattern, links to relevant articles (through Pubmed IDs), and summaries of the variant’s effect. Most importantly, scores can be entered for the variant in a series of categories related to evidence and clinical effect. These scores allow for the automatic sorting and filtering of variants -- once entered, a variant is considered “sufficiently evaluated” and can be used to automatically produce genome reports.
In keeping with the public sharing of genome and trait data, variant interpretations in GET-Evidence are freely shared as public domain under a CC0 license. GET-Evidence is a “peer production” model where all users are able to edit variants -- by allowing others to edit, mistakes can be easily corrected, updates in understanding based on new literature can be applied more rapidly, and consensus can form as multiple editors combine their knowledge and perspectives.
A public resource
We’re thrilled to have this paper published, formally introducing the PGP as a resource for researchers. We believe publicly shared data are invaluable for research and a key component of the scientific method. We also hope that GET-Evidence and our experiences with genome interpretation help others in the development of methods for genome interpretation. In publicly sharing data, the PGP has adopted a bold new method for human subjects research: an educated cohort consenting to the unforeseeable risks involved and a highly participatory ongoing relationship. A big thanks goes out not only to the coauthors on this paper but also to our many participants, for making this dream of a public resource a reality.
Since its inception the Personal Genome Project has pioneered new ways of doing research. The PGP and affiliated research groups remain focused on high-impact strategies for conducting research and for supporting and funding innovation. In particular, the PGP is considering ways to maximize the impact of personalized medicine and the use of whole genome sequence and related data to improve healthcare for all.
We’ve spoken to many PGP participants on a variety of topics and have heard many great ideas. Now we'd like your feedback on how we might work together on the broad areas of research described above--so we created the brief survey linked at the bottom of this post. We hope that responses from this survey encourage collaboration and grant funding for personalized genomic medicine research.
Although we're interested in building this collaborative infrastructure, we need to emphasize that the PGP does not have resources to support medical follow-up in response to genome findings. Even if we do receive some funding for personalized medicine research, it is likely to be quite limited. You should always assume that you'll be responsible for any and all medical care that may occur in response to research findings in our project. You can read more about this in Section 6.3 of our full consent for participation (Risks Associated with Your Receipt of Data From the PGP).
Please let us know what things you'd be interested in working on with us by filling out our survey:
Interest in Personalized Genomic Medicine Research (PGP blog survey, July 2012)
[youtube http://www.youtube.com/watch?v=BzoXJbRvPxs]
MIT Tech Review recently described Larry Smarr as the "Patient of the Future." As part of the morning Pioneer Session at the 2012 GET Conference, Larry gave an overview of his quantified self journey that led him to aid the diagnosis of a chronic condition. Watch the video.
[youtube http://www.youtube.com/watch?v=M4p3JI7s1dw]
As part of the Pioneer Session at the 2012 GET Conference, Eric Alm gave an overview of his effort to create a high resolution characterization of the microbial communities in his own gut by collecting and analyzing daily stool samples for a year (video).
I've heard Eric's research endearingly referred to as the "#2-ome"! Lawrence David, a former graduate student in Eric's lab at MIT, also participated in this self-study experiment. He runs an aptly named website: stinkpot.org.
http://www.youtube.com/watch?v=sJPJu9EEOAE&rel=0
George Church gave the opening remarks at the 2012 Genomes, Environments and Traits (GET) Conference on April 25th at Harvard Medical School (video). We'll be publishing more videos from the 2012 GET Conference soon.
Jason Bobe is Executive Director of PersonalGenomes.org, a 501(c)(3) that aims to make genomes useful for humankind.
Yesterday Madeleine Ball and I finished processing the 100+ microbiome kits that we collected at the 2012 GET Conference and shipped them on dry ice to our collaborators at the University of Colorado for analysis.
[caption id="attachment_605" align="aligncenter" width="577"] A (very pregnant!) Madeleine Ball processes microbiome kits and prepares them for shipment on dry ice to our collaborators at the University of Colorado. Photo Credit: Jason Bobe. CC-BY.[/caption]
A (very pregnant!) Madeleine Ball processes microbiome kits and prepares them for shipment on dry ice to our collaborators at the University of Colorado. Photo Credit: Jason Bobe. CC-BY.[/caption]
Each microbiome sampling kit contains 5 sterile swabs. PGP participants attending the GET Conference could volunteer to take a kit and swab 5 body sites: left hand, right hand, forehead, mouth and gut (via swabbing stool on a piece of used toilet paper -- yes, made for interesting conversation at the conference!). Out of the ~100 kits returned by PGP participants, only 1 kit excluded the gut sample; the remaining 99 kits had all 5 swabs. Our volunteers are amazing (and adventurous)!
The Knight Lab at the University of Colorado will perform 16S sequencing and analysis that will help us characterize the microbial diversity of these 500 samples. There are many interesting scientific questions that we will be able to begin to explore, such as: Do family members share similar microbes? What about unrelated persons sharing the same household? How do diet, medications, and various lifestyle factors affect microbial communities? Do these affects change with age? How do antibiotics affect our microbiomes? It will be really exciting to explore more deeply the linkages between microbes and certain diseases, such as skin infections, obesity, inflammatory bowel disease, etc.
Once the analysis is complete the results will be privately available to PGP participants. Then after 30 days, the data will become publicly available and associated with participant profiles. Stay tuned!
The 2012 GET Conference was generously sponsored by the DTC genetic testing company, 23andMe, who donated 10 coupons for free access to their Personal Genome Service. At last week's staff meeting, we randomly selected ten PGP participants who attended the 2012 GET Conference. Here are the winners:
hu2843C9
huEBD467
huED0F40
huCCAFD0
huA90CE6
huE9B698
huE58004
hu60AB7C
hu831FD6
huFFB09D
These 10 folks have 30 days from today to get in touch with me to claim their prize. Or, if you would prefer to donate your prize to another PGP participant, please let me know!
This is a guest post from Rob Stein, Correspondent/Senior Editor at National Public Radio (NPR).
I am a reporter at NPR. I am working on a story about whole genome sequencing. I'm trying to find people who have had their genomes sequenced to interview for my story. Ideally. I'd like to find some people who are not involved in genetics professionally, but had their genomes sequenced primarily out of personal curiosity. Please contact me at: 202-513-2794 or rstein@npr.org.
One thing the Personal Genome Project prides itself on is its ethics: the methods of "open consent" stem from seeking an ethical solution to the dilemmas presented by human genome research. Making your personal genome and health data public is still a radical departure from traditional research, and so it is important to us that participants truly understand the expectations and risks of participation. Indeed, the first ten participants (the PGP-10) were selected from individuals who had a master's level degree or equivalent training in genetics. Since then, participation has been opened to almost anyone1 -- provided they demonstrate an understanding of the expectations and risks involved in participation. As part of demonstrating this, prospective participants are asked to complete an "enrollment exam" testing their understanding of basic genetics concepts, human subjects research, PGP protocols, and some of the open-ended risks associated with participation.
Issues with the old exam
The exam experience has been unfortunately difficult for many, it is the single largest hurdle to participation. An analysis of user logs showed me that for every person that fully enrolled in the project, there was one that had given up at the enrollment exam. It wasn't meant to be exclusive, we welcome autodidacts: anyone meeting our minimum requirements1 should be able to join, provided they are willing to learn. Seeking to improve the experience, a couple months ago I decided to look at the user logs for the enrollment exam. What questions were problematic? Why was the exam so painful?
[caption id="attachment_478" align="aligncenter" width="498"] Histogram of PGP enrollment progress, logs from 12/2010 to 12/2011. Nearly as many people gave up at the enrollment exam as made it through to enrollment application submission & participation.[/caption]
Histogram of PGP enrollment progress, logs from 12/2010 to 12/2011. Nearly as many people gave up at the enrollment exam as made it through to enrollment application submission & participation.[/caption]
One problem was the design. Each module of the exam requires all questions to be answered correctly, and the user is not informed which was incorrect. Because one module contained nine questions, that module was a combinatorial nightmare! 10% of participants retook the module over a dozen times (and note: I was only looking at data from enrolled participants -- people who made it through the whole thing!).
[caption id="" align="aligncenter" width="500"] One in ten participants had to take the "Risks and Benefits" module at least a dozen times to pass the exam! This module had the most questions of any (nine), which was probably part of the problem.[/caption]
One in ten participants had to take the "Risks and Benefits" module at least a dozen times to pass the exam! This module had the most questions of any (nine), which was probably part of the problem.[/caption]
So one thing I changed in our updated exam was to reduce the number of questions in a single module: now the highest is six, and most modules have four. Another issue was a little less obvious, but seemed to be an issue based on user behavior and communications: it was unclear for many users how they could learn the correct answers to the questions. An old study guide was linked, but the link wasn't obvious (it appeared in the lower right on exam pages, while taking the test) and the site was difficult for some to navigate.
An updated exam with integrated study guide
To figure out which questions were problematic (or just plain out-of-date), I asked the PGP staff to go through the old exam and rate each question on relevance and clarity. Using this survey data, we hashed out which questions we wanted at one of our weekly meetings. Based on this, I went through and made a new exam: one with an integrated study guide for each module. All questions on this new exam are directly related to the study material, and the study guides can be revisited during the test. Hopefully the new design avoids the frustrating user experience of numerous repeated attempts, trying to find the correct answer combinations.
I hope the new exam is both easier and more educational. The PGP staff reviewed and tweaked the new exam and study material, then we submitted our updated exam for approval with our institutional review board at the start of April, it was approved April 30th, and Ward implemented the changes on the enrollment website last week. A document containing the study guides is available on the Harvard PGP website. It's not just for new recruits: if you're a current participant -- or even a curious onlooker -- reading this study material is a great way to learn more about the PGP!
1PGP minimum requirements include an age requirement (21 years or older) and US citizenship or permanent residency. This second requirement is because legal issues associated with human genome data are changing rapidly; our project is only prepared to handle the regulatory issues for the United States in particular. We hope PGP projects in other countries will grow and provide additional options internationally.
Daniel Vorhaus is an ethical and legal advisor to the Personal Genome Project, Editor of the Genomics Law Report and an attorney with Robinson Bradshaw & Hinson.
More than six years ago, I read a piece in Scientific American written by George Church entitled “Genomes for ALL” (pdf). Most of the article was devoted to the rapid progress of next-generation DNA sequencing technologies. But on the next to last page was a brief description of the Personal Genome Project, an audacious new project that aimed to recruit volunteers to provide genomic and phenotypic data and, then, to do something unexpected with those data: publish them for all the world to examine.
I was inspired by the project’s audacity. I was also a law school student with time on his hands. I immediately wrote to George and offered to help.
Fast forward six plus years to April 2012 and the Personal Genome Project’s third annual GET Conference. In the intervening time, the PGP had grown from one participant (George) to well over 1,000 enrolled and active participants, more than 100 of whom attended the GET Conference in person. And after years of revising enrollment exams and consent forms, negotiating IRB reviews and sponsorships and developing sample collection protocols and project management software, the Personal Genome Project had exploded off of the page and into the hallways of Harvard Medical School.
At one point in the middle of the conference I found myself surrounded, quite literally and for the first time, by the PGP. As all of the PGP participants and staff gathered together in a Harvard Medical School courtyard for the group photo*, I stopped to consider the situation: in less than half the time it took to sequence the first human genome, George’s audacious idea – the Personal Genome Project – was an idea no longer. This was the photographic proof. I was inspired all over again.
[caption id="attachment_469" align="aligncenter" width="600"] April 25, 2012. PGP participants at the 2012 GET Conference in Boston MA. * Photo credit: Mike Dravis and PersonalGenomes.org, CC-BY-SA[/caption]
April 25, 2012. PGP participants at the 2012 GET Conference in Boston MA. * Photo credit: Mike Dravis and PersonalGenomes.org, CC-BY-SA[/caption]
Much more than the photo above, the frenetic pace of the conference and the myriad activities for PGP participants attending in person attests both to how far the project has come, as well as to how many exciting developments wait just around the corner. Last week, in the space of mere hours PGP participants were invited to:
Many participants availed themselves of all of the above activities, and in the weeks and months to come the PGP will be returning more data (genomic, microbiomic, phenotypic) to more participants than ever before.
As we look forward from this year’s wonderfully successful GET Conference, we want to set our sights even higher for next year. Our goal for the 2013 GET Conference is to have even more PGP participants attending, and to see them engaged in an even broader array of the public and participatory research efforts that make the PGP unique.
Getting to that point will take lots of work, and we would welcome your help. If you are a PGP participant, please contact us and share your ideas for next year’s GET Conference (to be held April 25th and 26th in Boston, MA). Whether or not you’re a PGP participant, you can also help us to build on the momentum of this year’s GET Conference by volunteering or by donating.
With your help, we can continue to accelerate the work of turning this audacious idea into reality!
We're happy to announce that the Personal Genome Project has received its first donated 23andme exome from a participant! As with the genotyping data acquired from direct to consumer testing companies, the PGP also welcomes donations of larger data sets like genomes and exomes. When assigning PGP nicknames (like "PGP1"), we have decided they should go to individuals who have exome or genome data hosted in the PGP -- whether sequenced by us, or donated to us. Thus, hu97DB4A now has the nickname "PGP18"!
What is exome data?
In my earlier post "The Whole Two Yards" I explained that the PGP is interested in whole genomes rather than genotyping. Exome sequencing is a third category of DNA analysis. So what is an exome?
"Exome sequencing" refers to something much like genome sequencing, but limited to the regions in the genome that code for proteins (the "exons" of genes). Proteins perform most of the functions in the cell: from structures to enzymes to relaying signals, proteins are the workhorses of biology. Thus, most known genetic variations that have significant effects are the result of changes within exons -- changes that disrupt the resulting protein coded by a gene. Surprisingly, these regions only account for 1% of the genome! Because of this, there has been some focus on targeted sequencing of only exons -- hopefully getting almost as much useful sequence for a fraction of the cost.
[caption id="attachment_433" align="aligncenter" width="480"]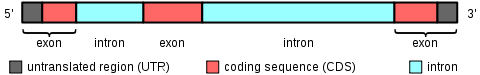 Genes contain protein coding regions ("exons") interspersed with large non-coding gaps ("introns"). To save money, exome sequencing targets and sequences only the exons in the genome -- thereby focusing on the regions most likely to have variations that affect traits. [Image by User:Daycd on en.wikipedia.org, shared as CC-BY-SA][/caption]In the end, isolating exons is difficult, so "exomes" aren't that much cheaper than whole genomes (maybe 2-3x cheaper, not 100x)... but it's still useful to have the cheaper option.1 23andme recently started a pilot exome sequencing service (notably, they provide no interpretation of the data), and some PGP participants have signed up for it.
Genes contain protein coding regions ("exons") interspersed with large non-coding gaps ("introns"). To save money, exome sequencing targets and sequences only the exons in the genome -- thereby focusing on the regions most likely to have variations that affect traits. [Image by User:Daycd on en.wikipedia.org, shared as CC-BY-SA][/caption]In the end, isolating exons is difficult, so "exomes" aren't that much cheaper than whole genomes (maybe 2-3x cheaper, not 100x)... but it's still useful to have the cheaper option.1 23andme recently started a pilot exome sequencing service (notably, they provide no interpretation of the data), and some PGP participants have signed up for it.
Addition of VCF interpretation in GET-Evidence
23andme provided the participant with both a VCF file and individual read data (in the form of a "BAM" file). Personally I'm not a fan of the VCF format for personal genomes, mainly because it fails to report which regions are confidently called as "matching reference". (What this means is that, if a variant isn't listed in the file, you can't tell whether (a) you don't have it, or (b) that region simply wasn't well covered.)
That said, VCF is a very common format, and so I've finally added the ability to interpret VCF to GET-Evidence. I ran the exome data through GET-Evidence and did a little bit of additional interpretation (as with other whole genome reports, these interpretation is far from complete). You can visit the report on GET-Evidence -- and if you'd like a copy of the VCF file itself, it is linked at the top of the report as "source data". We're hoping to reprocess the BAM files to produce higher quality reports and publicly host these larger files as well. For now, though, we're able to immediately accept and interpret VCF files.
Donation of genetic data is very valuable to our project, hopefully we'll see other 23andme exomes donated from participants in the future!
1Exomes have other issues that makes them less desirable, including extremely high variations in coverage, and are difficult to use for detection of larger structural variations (like large deletions or duplications of regions). The PGP does whole genome sequencing because we wish to collect the best data possible, and we feel that a full genome's data is worth the 2-3x higher cost.
Many thanks to all who participated, supported, and attended the GET2012 conference! Don't consider this to be our only follow-up post about the conference, I just wanted to write a brief message and share some photos of the sample collections. (One thing we'll post later is some videos online.)
We managed to collect blood from 110 participants and 80+ microbiome kits (we haven't counted them yet) -- it's really exciting to have so many samples! The blood will be especially valuable, many thanks to our colleagues at the Feinstein Institute (the nurses who did the blood draws). Anu, Tom, and Ward all helped pack the samples up for overnight shipping to Coriell to establish blood cell lines (EBV-transformed B cells, to be shared in the NIGMS repository). Here's Tom's parting photo of the kits, as he and Anu left them at FedEx while I circled the block in a Zipcar truck:
[caption id="attachment_401" align="aligncenter" width="600" caption="GET2012 blood kits at the FedEx office for overnight shipping. By Tom Clegg, shared as CC0."] [/caption]
[/caption]
And here's a snapshot we made on Monday evening, when Jeantine, Jason and I finished constructing the microbiome kits for the conference (five self-collected swabs from various body sites). Many thanks also go to John Aach & Mike Chou for their invaluable help, spending the better part of the day at the reception desk handing out and receiving these kits. An avalanche of ziploc bags!
[caption id="attachment_397" align="aligncenter" width="600" caption="Microbiome kits for GET2012 conference. By Madeleine Price Ball, shared as CC-BY."] [/caption]
[/caption]
Since PGP participants are geographically diverse, our sample collection methods often involve mailing sample collection kits to participants. Participants are asked to then claim the kit they receive as belonging to them (which kit is mailed to each participant is random, not assigned). When participants claim kits, they use the "kit nickname". Kit nicknames were also created to facilitate kit assembly. It is very difficult to check that a random ID string matches for all the tubes or swabs packed into a kit -- it's much easier to check that the kit nicknames all agree. Every time I assemble kits I find myself thankful for this innovation.
A random selection of nicknames that have already been used: "Brainard", "Patterson", "Crestway", and "Thetford". Kit nicknames aren't published on the website, we consider them semi-private: an added layer distancing the publicly shared kit ID from a participant's name and private identity.
Where did these names come from? Boston street names! If you check Google Maps you'll find that Boston contains "Brainard Street", "Patterson Way", "Crestway Road" and "Thetford Avenue". These came from public map data shared by the state of Massachusetts. I got the names thanks to my friend, Chris Schmidt, who has worked closely with open mapping projects and software over the years.1 I asked him about it thinking that street names tend to be fairly recognizable, readable & writable, and were unlikely to contain anything considered "inappropriate".
Boston gave us a little over 3,000 names and we were running low, so I revisited the page and downloaded data for all of Massachusetts, getting over 17,500 names (~14,500 of which were new). Afterwards, I looked at the names a bit and noticed something funny -- there's definitely some spelling errors in there, I noticed "Meadowowbrook Avenue" and "Jenniffer Road". It looks like the public data isn't perfect! The nicknames are all in the system, spelling errors included, so... if you receive a kit with something that looks like a spelling error, don't try to correct the spelling when claiming it. Just type it exactly as you see it, even if it looks funny!
1To generate the Boston kit nicknames, I ran the following (per Chris's instructions):
ogrinfo eotroads_35.shp -al | grep STREET_NAM | sort | uniq > streets.txt
This is a guest post from Andrew Evans, CEO of Genomic Arts and Senior Information Architect at 5AM Solutions. His company is sponsoring the 2012 GET Conference, where he'll be showing off "genetic avatar technology" for attendees. Here is a mini press release with more details.
Putting the "Fun" Back Into Functional Genomics
Rockville, MD-based Genomic Arts aims to put a friendlier face on personal genomics - literally. "Engaging the public in genetics research is critical to the advancement of precision medicine," said Genomic Arts CEO Andrew Evans. "While many people grasp the importance of it, they are often reluctant to participate - Genomic Arts seeks to make genomics fun and accessible to everyone."
As a hint of things to come, Genomic Arts is debuting its patent-pending genetic avatar technology at the 2012 Genomes Environments Traits (GET) Conference to be held at Harvard Medical School on April 25.
"Basically, we create a video game avatar character by reading your genetic code," Evans said. The technology demo, dubbed "23andMii" by Genomic Arts, currently reads single-nucleotide polymorphism (SNP) data created by the 23andMe personal genomics service, and g enerates an avatar in the style of a Nintendo Wii Mii character. "While this demo uses 23andMe and Nintendo platforms, the core technology is not limited to either one," Evans said.

Special Invitation
Genomic Arts is offering all GET attendees who have their own 23andMe genetic data the opportunity to have their avatar printed on their conference badges. "We know we won't get everyone's avatars right on the first try - this year's GET focus is on traits, and we hope this will spark attendees to discuss the strengths and weaknesses of calling phenotype directly from genotype. We will also ask for feedback from attendees about what they feel our software got right - and wrong - when generating their avatars," Evans said.
In addition, Genomic Arts will have a large display in the conference center featuring the avatars of all participating conference attendees.
GET attendees wishing to have genetic avatars printed on their badges will need to provide their 23andMe data to Genomic Arts by no later than April 20. To participate, please contact Andrew Evans at andrew@genomicarts.com
PGP participants - if you have already uploaded your 23andMe data file to your PGP profile, you may simply send along a link to your profile, or your profile ID.
#####
Genomic Arts. Genomic Arts, LLC is advancing genetics research by engaging the public in innovative ways. Genomic Arts was founded by bioinformaticians Andrew Evans and Will FitzHugh with the backing of Maryland-based life sciences software powerhouse 5AM Solutions. Contact: Andrew Evans - andrew@genomicarts.com
GET Conference. The GET Conference is the event for people working at the frontiers of human biology. We invite leading thinkers to discuss the important ways in which new genomic technologies will affect all of our lives in the coming years and to debate their technical, commercial, and societal impacts. We bring together scientists, industry leaders, entrepreneurs, practitioners, investors, researchers, and others to discuss advances in our ability to measure and understand human biology. The GET Conference is organized by the nonprofit charity PersonalGenomes.org. (Twitter: #GET2012)
As we produce and release participant genome data publicly, the Personal Genome Project also tries to provide some rudimentary interpretations of that genome data to participants (along with access to the genome sequence data itself). GET-Evidence is an open, collaborative system for genetic variant interpretation that has been developed to address this need. The database of variant interpretations is shared without restriction using CC0, and the associated software under a free software license.
I spent some time this last week adding guides to help people interested in the exploring the GET-Evidence system. Tom Clegg and I also did some brainstorming and added this summary image (below) to the front page. If you're interested in seeing some interpretations for PGP participants who have already had their genomes sequenced, check out GET-Evidence's genome reports page.
I’ve seen a number of participants write us emails asking: “Where’s my genome?” Believe me, we’re working on it! What we’re trying to do is pretty big, though -- and Rome wasn’t built in a day.
If you’re familiar with services like 23andme or FamilyTreeDNA, you might not realize that there are important differences between these “direct to consumer genetic testing” companies and the Personal Genome Project. One major difference is the technology being used. These companies perform genome-scale genotyping: a technology which analyzes a subset of genome positions known to vary between people. In contrast, the Personal Genome Project is interested in performing “whole genome sequencing” to gather information for all positions in the genome.
Each technology has some benefits and disadvantages...
| Genome-scale genotyping | Whole genome sequencing |
| 0.03% of the genome | 90-100% of the genome |
| Known variations (usually common) | All variations (common and rare, known and unexpected) |
| Ideal for ancestry, usefulness for health & traits is debated |
Ideal for both ancestry and health |
| Interpretation is facilitated -- there are already many published studies for known variations |
Interpretation is currently very difficult for rare and unexpected variations |
| $200-$3001 | $4,000-$5,0002 |
| Weeks to produce | Months to produce |
| 5-10MB of data | 40-400GB of data |
Currently, genome-scale genotyping has an excellent cost-benefit ratio. There’s a good reason it’s so popular -- whole genome sequencing simply costs too much! But the whole genome is 6 billion positions, any of which could potentially hold a rare and uniquely personal variation. If you stretch a single human genome’s DNA out like a string, this is a bit more than 2 yards in length (or 2 meters for the metric folks). In comparison, a molecule with 2 million positions (the number of positions typically genotyped) would only be about 1/40th of an inch (or 2/3 of a millimeter)!
The Personal Genome Project is dedicated to sequencing the whole two yards of each genome: the rare and the unique along with the common variations. We think it’s likely that analyzing only the common points of variation is going to miss out on the most dramatic, rare variations that affect our health and traits.
Whole genome sequencing technology is new. Rapid improvements are still being made, but it still takes a lot more of both time and money to get a whole genome. Right now it takes a couple of months to produce a genome, and it costs thousands of dollars. It’s expensive, but the price has finally decreased to a point where we decided it was time to get started.3 And so we are sequencing genomes.
Those of you who have been watching our project for a while might already know this... it literally took years for the first ten PGP participants (known as the “PGP-10”) to get their genomes after signing up and donating samples. We’re at an exciting point right now as we expand the project, but we can’t go from 10 to 1,000 in the blink of an eye!
Sample collection was one of the large barriers we had to overcome as we moved beyond the PGP-10. With the help of Pete Estep we now have saliva samples from nearly 1,000 people. Most participants who sent us samples won’t receive their whole genomes for a while yet, but it’s important that we have them. 200 genomes are currently queued up for sequencing in the coming months. Once we’ve done these we’ll have learned a lot about how to move forward, and we’ll be raising more funding to sequence our other samples.
Rather than receiving a polished service using existing published research, Personal Genome Project participants are on the cutting edge of creating new research discoveries. Even the way our study is organized (in particular, releasing personal genetic data publicly) is unprecedented in many ways. Working at the “cutting edge” of research is the opposite of “polished”: we’re venturing into uncharted territory. There’s a lot of unexpected decisions to make, trial and error, and false leads. It’s exciting stuff -- but sometimes it’s also frustrating! Many thanks to the individuals who have chosen to work together with us in this pioneering research.
Footnotes
1 Links: Info on pricing for 23andme and FamilyTreeDNA.
2 Links: Info on pricing for Illumina and Complete Genomics.
3 In the early days of the PGP, the cost of a whole genome sequence was as expensive as buying a nice house or a yacht!
Recently I've been working on making kits for collecting blood samples from participants. Here's a quick snapshot of some of the materials:
[caption id="attachment_274" align="aligncenter" width="600" caption="Various boxes for shipping the kits. On the bottom shelf are the boxes that ship to the Coriell Institute, for establishing cell lines. I've pre-labeled them with hazard labels to minimize packaging work for the phlebotomist. On top are some boxes from Infekta. Scary name, right? We didn't pick it -- it's a company that specializes in packaging materials for potential biohazards like blood. (Photo by Madeleine Ball, CC-BY-SA)"] [/caption]
[/caption]
Sample collection for the PGP is uniquely complicated. Most studies collect from people who are already patients, and they're all visiting a particular clinic -- but we want to collect nationwide. (No, you shouldn't have to fly out to Boston!) Our colleagues at the Feinstein Institute are working together with us, we hope we can create a kit that a network of phlebotomists can use. It has several blood collection tubes, packaging materials, overnight FedEx labels, and instructions for both participants and phlebotomists... there's a lot of moving parts! I've got a fair amount of experience programming computers, but these days it seems I'm programming humans...
Four brave participants in the New York City area have volunteered their time to pilot these with our Feinstein Institute colleagues (who are based in Manhasset, NY). Our fingers are crossed, it's exciting to develop a standard procedure for this!
The premise of the PGP is that matched sets of genome, environment, and trait information should be available to all investigators. Not just professional researchers with million dollar labs, but PGP participants themselves, family members, hobbyists, and students. Getting the data should be easy and free.
To this end, as we grow our database and our collection of tissue samples, we are also developing suitable publication mechanisms. One recent development is the directory of participants, at https://my.personalgenomes.org/users. This page lists all enrolled participants, and allows some basic sorting based on what kinds of data are available on their public profile pages.
Here are some things you might like to try:
Here are some things you can see on a public profile page, in case you haven’t noticed:
Our public data access tools will continue to grow as we accumulate participants, whole genome sequences, health records, and other data.
Finally, if you’re a machine, you can find the same information in a more readable form at the following address: https://my.personalgenomes.org/users?format=json&iDisplayStart=0&iDisplayLength=15.
The other day someone I work with closely called in sick (or rather, "emailed in") with a case of stomach flu, and expressed a hope that I was not infected. I was fine ... in fact ... I don't recall ever having a stomach flu. A colleague in David Reich's lab had mentioned that resistance to stomach flu is fairly common, and is tested by 23andme. Do I have this resistance? I checked, and I do! What's the causal variation? How does it work?
The genetic variation that makes me resistant is actually related to the ABO blood group system. ABO blood groups are a set of modifications made to the proteins that coat our red blood cells. The default modification is "O", while people with A or B genes can also have "A" and "B" modifications. In most people these modifications are also expressed on the surface of other cell types, including cells that line our gut and mouth.
Two genes are involved in placing the modifications on different cell types: FUT1 for red blood cells, and FUT2 for other cells ("mucosal tissues" and saliva). There is a common genetic variant that breaks the gene FUT2 by creating a stop codon (variously identified as rs601338, FUT2-W143X, and FUT2-W154X). 20% of Europeans and Africans have this highly disruptive variation on both copies of the gene (from both mom and dad). These people are "non-secretors": the ABO modifications don't occur on the surface of cells in their gut tissue.
[caption id="attachment_232" align="aligncenter" width="468" caption="ABO blood types involve modifications made to proteins that coat the surface of blood cells. Usually these modifications also appear on other cell types (including cells lining the gut and stomach) -- but 20% of Africans and Europeans are "non-secretors", due to a variant breaking the gene FUT2. (Image created by Madeleine Price Ball, shared under CC-BY-SA.)"]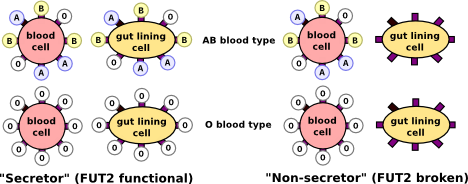 [/caption]
[/caption]
Most cases of stomach flu are caused by strains of "norovirus" (formerly called "Norwalk virus", often seen in cruise ship outbreaks). For norovirus to infect you, it needs to invade your cells -- and to do this, it needs to detect and attach to the cell's surface. Most noroviruses attach to the ABO modifications on the surface of gut tissue cells, using this to invade. Viruses generally recognize all three types pretty well (A, B, or O...although there's some reports of biases) ... more importantly, if none of these are present the virus can't invade! Non-secretors are extremely resistant to most strains of norovirus.
[caption id="attachment_211" align="aligncenter" width="519" caption="Secretors express their ABO blood group modifications on the surface of gut tissue cells. Norovirus uses these to recognize and enter the cells. As a result, non-secretors (who don't have these modifications in gut tissue) are extremely resistant to the virus -- the virus can't find the cell, and thus can't infect it. (Image created by Madeleine Price Ball, shared under CC-BY-SA.)"]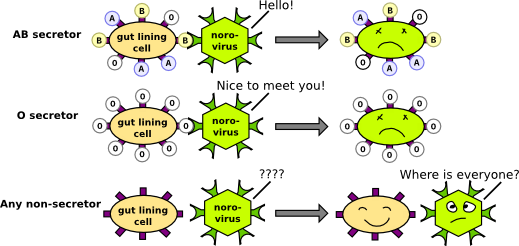 [/caption]
[/caption]
There are still some strains of norovirus that will get me sick, so while I can say that I am "norovirus resistant", I would be stretching it to say I'm "invulnerable". These strains have evolved to recognize something different, instead of ABO modifications. That said, it's certainly a a nice trait to have, and one most people who have it wouldn't realize -- how many of us notice not getting sick?
Although a mutation destroying a gene is usually a bad thing, in this case it's beneficial -- it's actually a variant that protects against disease. As part of our automatic interpretations of genomes I've added a review of this variant to GET-Evidence, our public database of variant interpretations. It now shows up on genome reports; so far I can see that two members of the PGP-10 (PGP6 and PGP7) share this secret superpower with me.
Preston Estep, PhD is the Director of Gerontology and leads the saliva collection effort at the Personal Genome Project at Harvard Medical School.
We’re getting to love spit. Here are a couple of fun facts about saliva that most people (including most scientists) don’t know: most saliva is loaded with DNA, and the primary source of that DNA is blood cells. This has been shown by genotyping people with bone marrow transplants (1). The genotype of blood and saliva from transplant recipients is typically more similar to the genotype of the marrow donor than the pre-transplant genotype of the recipient (the genotype of the non-hematopoietic cells of the recipient is retained). This amazing discovery also demonstrates interesting and practical things about saliva biology, including that most DNA in it does not come from cheek or other epithelial cells. So, when we send you a collection kit there is no need to swab or abrade cheeks (or to make awful choking sounds). Just salivate and spit.
The blood origin of saliva DNA is just one reason we love spit. And we need spit. Your spit! We know most of you have sent us saliva but we need those who haven’t to spit our way. Not in our general direction, but according to our directions, which can be found on a double-sided sheet of paper included in every kit we send out. For those who have provided saliva in the recent past, fear not, you’ll get yet another chance to spit again very soon. Plus, you’ll get to use our new and improved collection kits. We understand if you can barely contain your excitement.
If you are wondering about the status of saliva samples you already submitted, you can check your sample log. To do this, follow these steps:
[caption id="attachment_170" align="aligncenter" width="575" caption="Saliva samples donated by PGP participants in the freezer. By Preston Estep, licensed under CC-BY-SA."] [/caption]
[/caption]
If we have received your samples then they are in there somewhere. This one freezer is pretty empty and we have more to fill. We’re looking forward to you spitting our way.
References:
1) Thiede C, Prange-Krex G, Freiberg-Richter J, Bornhäuser M, Ehninger G. Buccal swabs but not mouthwash samples can be used to obtain pretransplant DNA fingerprints from recipients of allogeneic bone marrow transplants. Bone Marrow Transplant. 2000 Mar;25(5):575-7. PMID: 10713640
[caption id="attachment_92" align="alignright" width="160" caption="Joe Thakuria draws John Lauerman's blood for whole genome sequencing. By Madeleine Price Ball, licensed under CC-BY-SA."] [/caption]
[/caption]
Madeleine Price Ball, PhD is a PGP research scientist in George Church’s lab at Harvard Medical School.
Several months ago John Lauerman, a reporter for Bloomberg News, approached the Personal Genome Project interested in having his whole genome sequenced. While we have hundreds of genomes in the sequencing pipeline, of the dozen or so genomes we have sequenced to-date, so far the results have been for the most part uneventful.
Lauerman's case was different: we found something rare and "famous", and something that nobody could have anticipated by looking through family history: a mutation that was acquired rather than inherited. This genetic variant (JAK2-V617F) is one of a number of mutations that can accumulate in blood stem cells, a precursor that could lead to several rare blood diseases.
Last night Lauerman published his experience, and we encourage all participants to read it. It confronts us with a scenario that seems likely to affect others who forge into this new and unknown territory: the very real possibility that whole genome sequencing may uncover something unexpected, ambiguous, and scary. This certainly isn’t an outcome we anticipate for most participants, but it is a rare possibility all should be aware of. Would you rather know that you carry such a variant, even if that knowledge might not help your health at all? Although some would decline, PGP participants are the sort of people who say: "Yes, I’ll take that risk, I’d rather know!" [see footnote]
His experience also illustrates potential for the Personal Genome Project to guide health care, for himself and for those who follow. The JAK2-V617F variant is so rarely seen in healthy individuals, we have very little understanding of what to expect. It has almost always been seen after a patient is diagnosed with a disease, not before. Will he develop one of these diseases? If so, which one? Perhaps many people carry the variant but never develop any symptoms of disease. In coming years Lauerman will likely continue to monitor his blood for signs of disease. It is possible that he will never develop the disease, and we hope this is the case. On the other hand, through monitoring he may detect disease sooner than he otherwise would have. By making his experiences public, his case can inform future individuals who confront the same finding.
As we move onward to sequencing hundreds and thousands of genomes, we can’t promise such interpretations will be made in a timely manner. We're working with other groups to improve our ability to interpret genomes -- and PGP participants are the perfect testbed for this development! -- but it's much harder than you might think. Genome data is made public in 30 days, but months or even years could pass before a serious and potentially scary variant is noticed. Participating in the PGP not only means that you risk learning ambiguous and scary news, but that it may be uncovered long after your data has been made public. We are always grateful to participants who choose to step into that unknown territory of genome sequencing, and who share their data so that others may learn.
Footnote: In the early stages of enrollment, individuals interested in joining the Personal Genome Project are asked to think about whether there are specific types of genetic information that they might not want to learn about themselves. Our examples include medical conditions with no effective cures or therapies, cancer, degenerative diseases, and stigmatized traits (e.g. mental illness). We do not offer the review or redacting of such information on a case-by-case basis. Only participants who wish to take the risk of learning such information are allowed to proceed with enrollment.
Madeleine Price Ball, PhD is a PGP research scientist in George Church's lab at Harvard Medical School.
This is a personal post, my recent story of personal genomic analysis. Although I am not a PGP participant, the story is an illustration of the personal empowerment that can be realized by having access to one's own genetic data.
The personal context for this news is that I'm pregnant, and two weeks ago I found out I was carrying a boy.
Normally such news would be interesting, but neither good nor bad. In my case, however, I was really hoping for a girl – for some concrete reasons of family history. I have two uncles (my mother's brothers) with mental retardation, likely caused by an unknown mutation. If that mutation is X-linked, there's a chance I'm a carrier for that X-linked mutation. If I have a boy, there's a chance I could pass it on to him.
Assuming it is X-linked, what are my chances of being a carrier? My mother had a 50% chance of inheriting it and I had a 50% chance of inheriting one from her – a total of 25% chance (50% of 50%). However I have something in my favor that makes this chance lower: I have two brothers who are clearly unaffected. Applying this new information to a Bayesian calculation, my mother's chance of being a carrier becomes 20% and my own becomes 10%.
I would have a 5% chance of passing it on to a male child (50% of 10%). Of course, whatever they have might not be X-linked (see footnote). I estimate the chances their issue is X-linked is around 50%, which would make the risk 2-3% instead of 5%. This is small but not trivial – it's like the risk a 44-year-old woman would have for Down Syndrome in her child. It is standard to offer such an expectant mother the option to test her fetus and choose to abort it (a common decision, if controversial). I have no such option, we can't test for a mutation if we don't know what it is.
And so, when I told my mother the sex, she decided to try to have her brothers diagnosed ASAP. It's not something she wanted to do, it would be difficult – it would require discussion with their caretakers, arrangements with medical providers, and it would disrupt their routine.
I was thinking hard about what I could do myself. My mind challenged with this new stressful information, I thought of something I could have done before now but hadn't occurred to me – I could use the genetic data from my family members to determine how much “at risk” X chromosome DNA I carry.
A map of the X inheritance in my family is below (apologies to colorblind people, I believe you can read “magenta, red, and cyan” as “blue, yellow/black, and gray”):
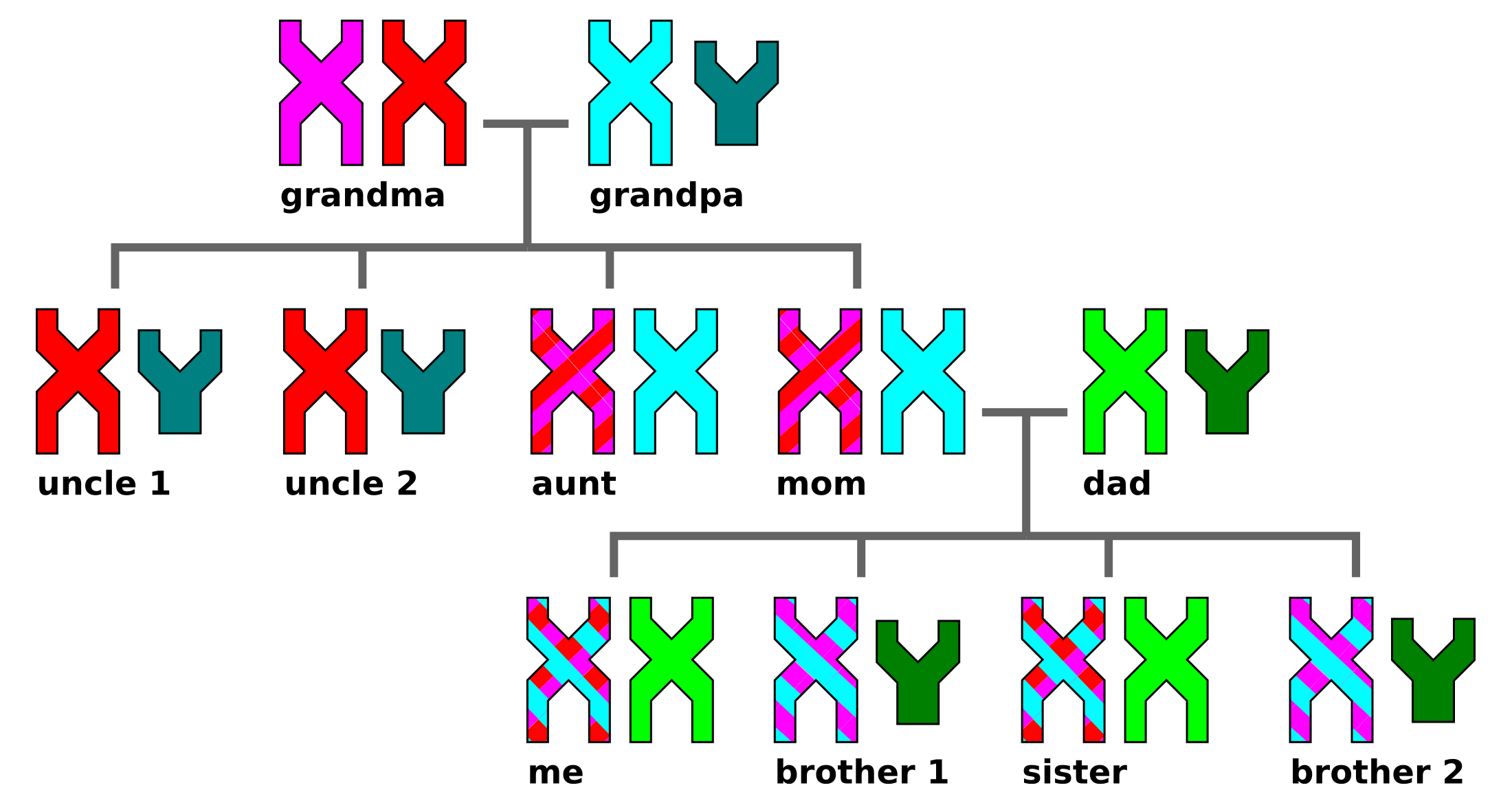
My grandmother's X's are represented as magenta (safe) and red (mutation bearing) – my aunt and mother are represented as inheriting a red/magenta mixture from her (we don't know which they got) and a cyan X from my grandfather. My sister and I inherited (1) an X from mom that is a mixture of my grandfather's cyan X and the unknown red/magenta from my grandmother, and (2) an X from our father. My brothers inherited a single X from my mother's parents – and here I've colored it as only magenta & cyan, because any parts they got from my grandmother are proven to be “safe” by the fact that they are not affected.
Such a diagram would lead you to think that inheritance blends the DNA up pretty well each generation, but it really doesn't – in reality inheritance is extremely blocky. 23andme provides diagrams of these blocks using the “Family Inheritance” and “Family Inheritance: Advanced” tools. Because my grandfather and brothers and I have all done 23andme, I can figure out which blocks I inherited from my grandmother and compare them to what my brothers inherited. If either of them inherited the same regions, then I knew those parts were confirmed “safe”. I only have to worry about regions I inherited from my grandmother and both brothers did not inherit.
Based on 23andme, I have the following maps for the X chromosomes of myself and my brothers. What I did was perform a comparison between each of us and our grandfather (all of whom have 23andme data); any regions matching my grandfather were marked as inherited from him, any remaining regions marked as from my grandmother.
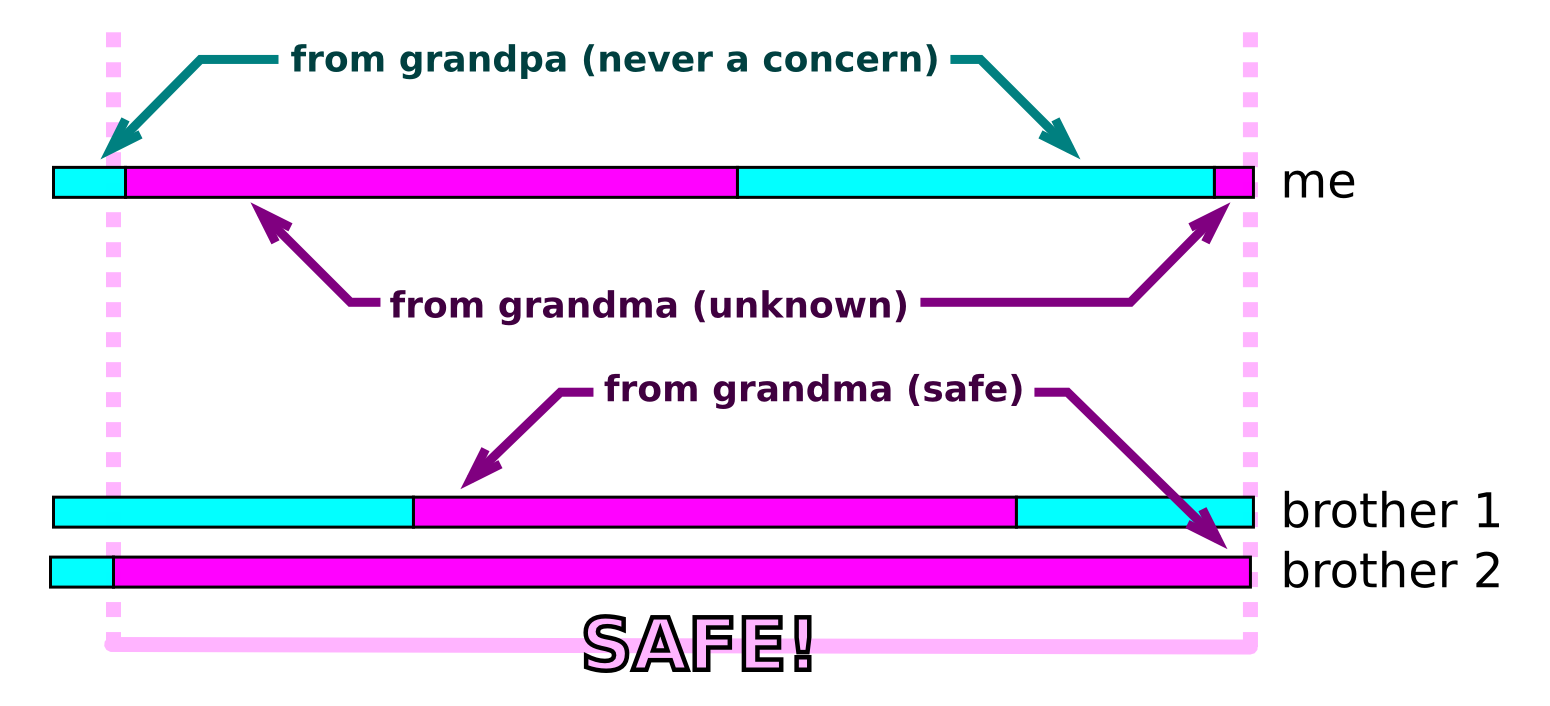
As you can see, it looks like I'm in the clear! Amazingly, brother 2 (his name is Andrew) managed to “test” a whopping 95% of the X our mother inherited from our grandmother! Thank you Andrew, excellent work! While there's 5% from our grandmother that neither brother has, I don't have it either so it's not an issue for me. Our sister does carry that segment -- some further data from my uncles would be useful for her (do they both carry that segment?), but there is no pressure to do it quickly.
Because diagnosis of my uncles was never certain, it's possible this has saved me years of uncertainty as I watched the child for signs of developmental delay. I'm extremely relieved -- and I'm really happy the "direct to consumer" genetic testing industry exists, allowing me to do this analysis. The result could easily have fallen in the other direction: if I found a larger-than-expected untested fraction from our grandmother then my risk estimates would have increased rather than disappeared. I am lucky it turned out the way it did. It's a happy ending for me, and now I can return to the mundane common concerns of pregnancy and children.
Note: this entry is adapted from a recent personal blog entry at http://www.madprime.org/.
Footnote: X-linked disorder was the major concern here. The other major possibility would be an “autosomal recessive” disorder, but the chances I would have an affected child even if I carried such a gene would be extremely low – Chris would also have to be a carrier for a mutation in the same gene (presumably an extremely rare event).
Other possibilities I could worry about carrying include Fragile X and balanced translocations. Fragile X is a string of trinucleotide repeats that can expand to a disease-causing length when passed from mother to child – I suppose it's theoretically possible one or both of my brothers and I inherited “premutation” versions and that mine could expand into “mutation” when I pass it to a child. It's also theoretically possible that my grandmother had a “balanced translocation” where a disease is caused if you only get one or the other, but not if you get both – in such a situation it's possible (but very unlikely) that my mother got both, somehow only passed zero or two to each of her children, and passed the same balanced set to me. I thought both of these were very unlikely, but I was tested for them – as I suspected, I carry neither.
Each year around the time of the "National DNA Day" holiday in late April, we invite leading thinkers to come together at the GET Conference to discuss the important ways in which new genomic technologies will affect all of our lives in the coming years and to debate their technical, commercial, and societal impacts.
We are excited that this year on April 25th, the GET Conference will be held on our home turf of Harvard Medical School. The Department of Genetics has co-sponsored the event and they are helping us bring Svante Pääbo to speak about his work on the Neandertal genome! We have many fabulous speakers lined-up, and I'll be sharing more details about them on this blog between now and April.
We are all very excited about something else that will be new at the GET Conference this year: we are adding a track to the event designed specifically for participants in the Personal Genome Project! We are thinking that this track may grow to become our annual "PGP users conference" and that we can use this forum to share progress reports and announcements about new initiatives from the PGP community. For those who are able to make the trip, we want to take advantage of the proximity as much as possible, so we are planning a some activities. For example, we are going to have an interactive area with a variety trait measurement and phenotyping booths. We will also have an area staffed with nurses where PGP participants may donate a blood sample or other specimens to be incorporated in the research study. We also hope this will be a time for participants, scientists, and others in the community to interact with each other.
Enrolled participants in the PGP may register to attend at no cost, for everyone else tickets are $225. If you are an enrolled participant in the PGP, please log-in to access your Promo Code. For PGP participants unable to travel to Boston, we will share videos of the event and find ways to keep you engaged from afar! More details soon!
Please visit the website: www.getconference.org
Here are a few names we came up with the for the blog. Which are your favorites?
[polldaddy poll=5918787]
Have other ideas? Leave them in the comments.