Part of what makes Personal Genome Project participant data uniquely valuable is our publicly shared trait data connected to public genetic data. A year ago our project was frustrated when our best resource for importing health data -- Google Health -- was discontinued. Ward soon got an interface running to import data from Microsoft HealthVault; the CCR-format data it produces is very similar, but isn't trivially combined with our Google Health records.1 We wanted to improve the quality of our trait data and provide another option for adding traits to public profiles.
And so we created a set of twelve trait surveys (the links below will only work for PGP participants) covering 239 traits and diseases:
To select what traits and conditions to include, the Google Health data was an invaluable resource. I was able to combine conditions using their ICD-9 codes (or, if unavailable, by internal Google codes).2 Here's the five most common reported traits:
[caption id="attachment_827" align="aligncenter" width="540"]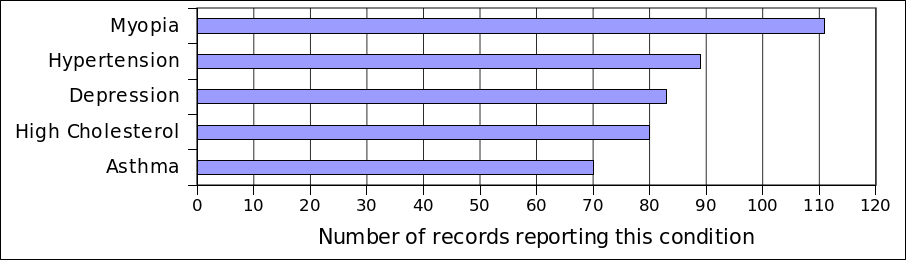 Top five conditions reported on Google Health records contributed by PGP participants.[/caption]
Top five conditions reported on Google Health records contributed by PGP participants.[/caption]
We tried to settle on four encodings corresponding to each trait: ICD-9, ICD-10, SNOMED CT, and NCIMetathesaurus CUI.3 I've shared our list of traits surveyed, along with the encodings we consider them associated with, as a Google spreadsheet.
A useful aspect of the ICD encodings is their organization by topic, and so our traits were split into twelve survey topics by ICD-9 encoding. It's impossible to be perfect in a first pass, but we tried to include anything that was fairly well-defined, not too rare (a prevalence of at least 1 in 10,000), and within the twelve ICD-9 ranges selected for the surveys. You might notice that some ICD-9 ranges were not used -- most notably, the category of mental traits and disorders. We do hope to survey these as well, but I want to be sure that participants are able to easily manipulate data on their public profiles before adding such a potentially sensitive category.
All PGP participants are invited to enter public trait data using these surveys -- although contributing such information is optional, and not required for participation. Even if you don't see a condition listed in the survey that you want to add, submitting an empty survey is useful information. I hope to follow up soon with a blog post analyzing some of the resulting data.
1On top of it, these records contain identifying data (like names and email addresses) that our participants weren't intending to make public. This meant we couldn't share the raw data, anything we shared was limited by our private CCR data interpretation process. Ideally we wouldn't be in this position: sharing raw data and allowing others to interpret it would be better, scientifically.
2If you're interested in it, this data was made available as "Dataset S1" in our recent open-access PNAS publication.
3Why four different coding systems? A couple of reasons: for redundancy, to facilitate using our data in other systems, to provide a starting point for harmonizing data from imported health records, and because we weren't (and still aren't) sure whether or how we'll be able to work with the licensing issues associated with some of these popular encoding systems.